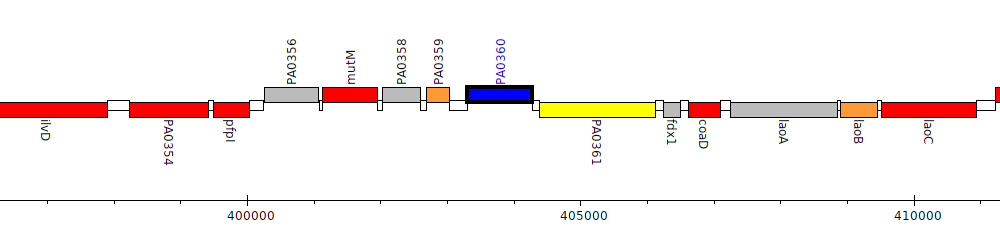
Pseudomonas aeruginosa PAO1, PA0360
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.25.40.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:1.25.40.10 | Tetratricopeptide repeat domain | IPR011990 | Tetratricopeptide-like helical domain superfamily | 17 | 193 | 1.2E-39 |
| PANTHER | PTHR43628 | ACTIVATOR OF C KINASE PROTEIN 1-RELATED | - | - | 52 | 167 | 1.7E-43 |
| SMART | SM00671 | sel1 | IPR006597 | Sel1-like repeat | 165 | 216 | 91.0 |
| Pfam | PF08238 | Sel1 repeat | IPR006597 | Sel1-like repeat | 129 | 164 | 2.1E-7 |
| Pfam | PF08238 | Sel1 repeat | IPR006597 | Sel1-like repeat | 201 | 216 | 0.022 |
| Gene3D | G3DSA:1.25.40.10 | Tetratricopeptide repeat domain | IPR011990 | Tetratricopeptide-like helical domain superfamily | 194 | 275 | 1.8E-13 |
| SMART | SM00671 | sel1 | IPR006597 | Sel1-like repeat | 61 | 92 | 20.0 |
| SUPERFAMILY | SSF81901 | HCP-like | - | - | 196 | 259 | 1.44E-11 |
| SMART | SM00671 | sel1 | IPR006597 | Sel1-like repeat | 217 | 252 | 2.8E-4 |
| Pfam | PF08238 | Sel1 repeat | IPR006597 | Sel1-like repeat | 94 | 128 | 1.2E-9 |
| SMART | SM00671 | sel1 | IPR006597 | Sel1-like repeat | 129 | 164 | 1.7E-7 |
| Pfam | PF08238 | Sel1 repeat | IPR006597 | Sel1-like repeat | 217 | 251 | 2.7E-4 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 290 | 328 | - |
| Pfam | PF08238 | Sel1 repeat | IPR006597 | Sel1-like repeat | 67 | 91 | 1.3 |
| SUPERFAMILY | SSF81901 | HCP-like | - | - | 52 | 177 | 2.49E-30 |
| SMART | SM00671 | sel1 | IPR006597 | Sel1-like repeat | 93 | 128 | 2.9E-10 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.