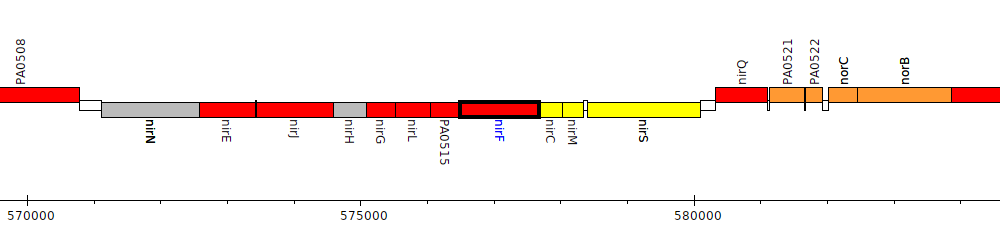
Pseudomonas aeruginosa PAO1, PA0516 (nirF)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006091 | generation of precursor metabolites and energy | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0051188 | obsolete cofactor biosynthetic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Biosynthesis of heme d1 |
ECO:0000037
not_recorded |
|||
| PseudoCAP | Denitrification |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR47197 | PROTEIN NIRF | - | - | 98 | 389 | 8.7E-33 |
| SUPERFAMILY | SSF51004 | C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase | IPR011048 | Cytochrome cd1-nitrite reductase-like, haem d1 domain superfamily | 26 | 390 | 3.66E-75 |
| Gene3D | G3DSA:2.140.10.20 | - | IPR003143 | Cytochrome cd1-nitrite reductase, C-terminal domain superfamily | 19 | 389 | 4.1E-49 |
| Pfam | PF02239 | Cytochrome D1 heme domain | - | - | 25 | 386 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.