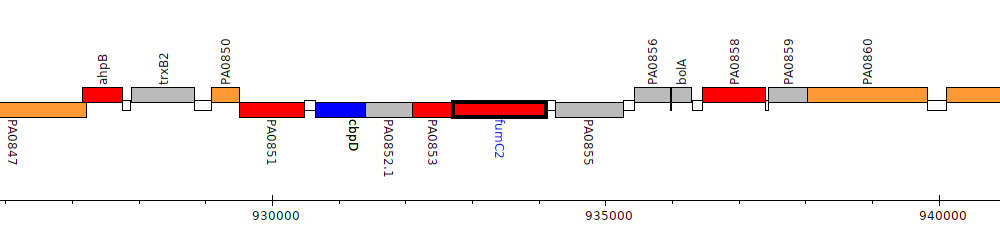
Pseudomonas aeruginosa PAO1, PA0854 (fumC2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0004333 | fumarate hydratase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd01362
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016829 | lyase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF10415
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF48557
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006106 | fumarate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:cd01362
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006099 | tricarboxylic acid cycle |
Inferred from Sequence Model
Term mapped from: InterPro:PF10415
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0045239 | tricarboxylic acid cycle enzyme complex |
Inferred from Sequence Model
Term mapped from: InterPro:cd01362
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCyc | TCA | TCA cycle I (prokaryotic) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Citrate cycle (TCA cycle) |
ECO:0000037
not_recorded |
|||
| PseudoCyc | GLYCOLYSIS-TCA-GLYOX-BYPASS | superpathway of glycolysis, pyruvate dehydrogenase, TCA, and glyoxylate bypass | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00620 | Pyruvate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | ANARESP1-PWY | anaerobic respiration | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01200 | Carbon metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | P22-PWY | acetyl-CoA assimilation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | TCA-GLYOX-BYPASS | superpathway of glyoxylate bypass and TCA | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | FERMENTATION-PWY | mixed acid fermentation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00020 | Citrate cycle (TCA cycle) | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| CDD | cd01362 | Fumarase_classII | IPR005677 | Fumarate hydratase, class II | 3 | 457 | 0.0 |
| PRINTS | PR00149 | Fumarate lyase superfamily signature | IPR000362 | Fumarate lyase family | 269 | 296 | 1.9E-32 |
| PRINTS | PR00145 | Argininosuccinate lyase family signature | - | - | 131 | 153 | 1.5E-6 |
| PANTHER | PTHR11444 | ASPARTATEAMMONIA/ARGININOSUCCINATE/ADENYLOSUCCINATE LYASE | IPR005677 | Fumarate hydratase, class II | 2 | 460 | 0.0 |
| FunFam | G3DSA:1.20.200.10:FF:000001 | Fumarate hydratase, mitochondrial | - | - | 137 | 404 | 1.7E-117 |
| Pfam | PF10415 | Fumarase C C-terminus | IPR018951 | Fumarase C, C-terminal | 406 | 458 | 3.5E-26 |
| PRINTS | PR00149 | Fumarate lyase superfamily signature | IPR000362 | Fumarate lyase family | 132 | 150 | 1.9E-32 |
| SUPERFAMILY | SSF48557 | L-aspartase-like | IPR008948 | L-Aspartase-like | 3 | 459 | 0.0 |
| FunFam | G3DSA:1.10.275.10:FF:000001 | Fumarate hydratase, mitochondrial | - | - | 3 | 136 | 1.1E-59 |
| NCBIfam | TIGR00979 | JCVI: class II fumarate hydratase | IPR005677 | Fumarate hydratase, class II | 3 | 458 | 0.0 |
| PRINTS | PR00145 | Argininosuccinate lyase family signature | - | - | 315 | 331 | 1.5E-6 |
| PRINTS | PR00145 | Argininosuccinate lyase family signature | - | - | 173 | 193 | 1.5E-6 |
| Gene3D | G3DSA:1.20.200.10 | Fumarase/aspartase (Central domain) | - | - | 137 | 403 | 3.0E-96 |
| Gene3D | G3DSA:1.10.40.30 | - | - | - | 406 | 462 | 7.9E-28 |
| Hamap | MF_00743 | Fumarate hydratase class II [fumC]. | IPR005677 | Fumarate hydratase, class II | 2 | 460 | 89.637482 |
| Pfam | PF00206 | Lyase | IPR022761 | Fumarate lyase, N-terminal | 10 | 340 | 2.6E-107 |
| PRINTS | PR00149 | Fumarate lyase superfamily signature | IPR000362 | Fumarate lyase family | 178 | 196 | 1.9E-32 |
| Gene3D | G3DSA:1.10.275.10 | - | IPR024083 | Fumarase/histidase, N-terminal | 3 | 136 | 8.9E-63 |
| FunFam | G3DSA:1.10.40.30:FF:000002 | Fumarate hydratase class II | - | - | 405 | 461 | 3.2E-29 |
| PRINTS | PR00149 | Fumarate lyase superfamily signature | IPR000362 | Fumarate lyase family | 315 | 331 | 1.9E-32 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.