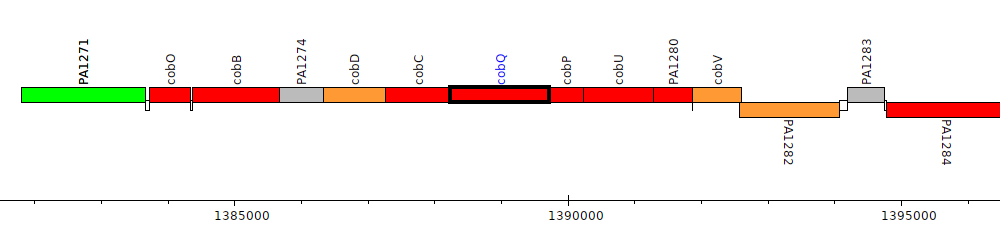
Pseudomonas aeruginosa PAO1, PA1277 (cobQ)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0051921 | adenosylcobyric acid synthase (glutamine-hydrolyzing) activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
1655697 | Reviewed by curator |
| Biological Process | GO:0009236 | cobalamin biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00028
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00028
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY-5508 | adenosylcobalamin biosynthesis from cobyrinate a,c-diamide II | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00860 | Porphyrin and chlorophyll metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY-5509 | adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | P381-PWY | adenosylcobalamin biosynthesis II (late cobalt incorporation) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCAP | Cobalamin biosynthesis |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 6 | 240 | 2.83E-58 |
| PANTHER | PTHR21343 | DETHIOBIOTIN SYNTHETASE | - | - | 6 | 486 | 0.0 |
| Gene3D | G3DSA:3.40.50.880 | - | IPR029062 | Class I glutamine amidotransferase-like | 257 | 401 | 7.5E-16 |
| Pfam | PF01656 | CobQ/CobB/MinD/ParA nucleotide binding domain | IPR002586 | CobQ/CobB/MinD/ParA nucleotide binding domain | 8 | 233 | 4.1E-23 |
| CDD | cd01750 | GATase1_CobQ | IPR033949 | Cobyric acid synthase, glutamine amidotransferase type 1 | 255 | 444 | 4.57272E-75 |
| SUPERFAMILY | SSF52317 | Class I glutamine amidotransferase-like | IPR029062 | Class I glutamine amidotransferase-like | 254 | 447 | 5.77E-42 |
| NCBIfam | TIGR00313 | JCVI: cobyric acid synthase CobQ | IPR004459 | Cobyric acid synthase CobQ | 8 | 478 | 0.0 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 3 | 244 | 5.6E-62 |
| CDD | cd05389 | CobQ_N | IPR047045 | Cobyric acid synthase, N-terminal domain | 6 | 228 | 0.0 |
| Hamap | MF_00028 | Cobyric acid synthase [cobQ]. | IPR004459 | Cobyric acid synthase CobQ | 1 | 488 | 216.30957 |
| Pfam | PF07685 | CobB/CobQ-like glutamine amidotransferase domain | IPR011698 | CobB/CobQ-like glutamine amidotransferase | 254 | 437 | 6.9E-58 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.