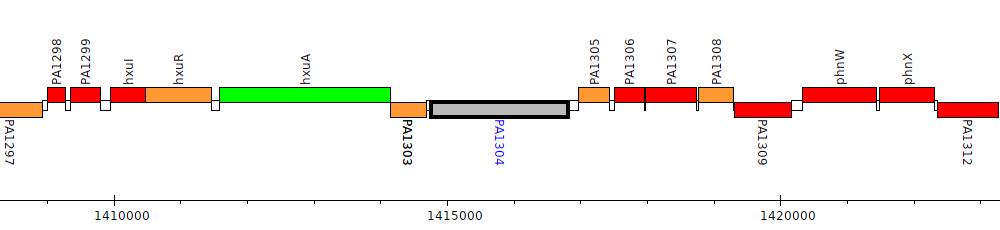
Pseudomonas aeruginosa PAO1, PA1304
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0008236 | serine-type peptidase activity |
Inferred from Sequence or Structural Similarity
Term mapped from: PDB:2XE4
|
ECO:0007090 |
31631799 | Reviewed by curator |
| Molecular Function | GO:0070012 | oligopeptidase activity |
Inferred from Sequence or Structural Similarity
Term mapped from: PDB:2XE4
|
ECO:0007090 |
31631799 | Reviewed by curator |
| Molecular Function | GO:0008236 | serine-type peptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00326
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PF00326
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF02897
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.40.50.1820:FF:000005 | Prolyl endopeptidase | - | - | 382 | 676 | 4.7E-94 |
| Gene3D | G3DSA:3.40.50.1820 | alpha/beta hydrolase | IPR029058 | Alpha/Beta hydrolase fold | 16 | 678 | 0.0 |
| PANTHER | PTHR11757 | PROTEASE FAMILY S9A OLIGOPEPTIDASE | - | - | 8 | 681 | 0.0 |
| SUPERFAMILY | SSF53474 | alpha/beta-Hydrolases | IPR029058 | Alpha/Beta hydrolase fold | 417 | 660 | 1.52E-42 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 504 | 523 | 1.7E-51 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 534 | 554 | 1.7E-51 |
| Gene3D | G3DSA:2.130.10.120 | - | - | - | 65 | 404 | 0.0 |
| SUPERFAMILY | SSF50993 | Peptidase/esterase 'gauge' domain | - | - | 2 | 410 | 1.88E-86 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 592 | 607 | 1.7E-51 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 450 | 468 | 1.7E-51 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 610 | 632 | 1.7E-51 |
| Pfam | PF02897 | Prolyl oligopeptidase, N-terminal beta-propeller domain | IPR023302 | Peptidase S9A, N-terminal domain | 12 | 402 | 8.6E-79 |
| Pfam | PF00326 | Prolyl oligopeptidase family | IPR001375 | Peptidase S9, prolyl oligopeptidase, catalytic domain | 466 | 681 | 9.3E-67 |
| PRINTS | PR00862 | Prolyl oligopeptidase serine protease (S9A) signature | IPR002470 | Peptidase S9A, prolyl oligopeptidase | 476 | 500 | 1.7E-51 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.