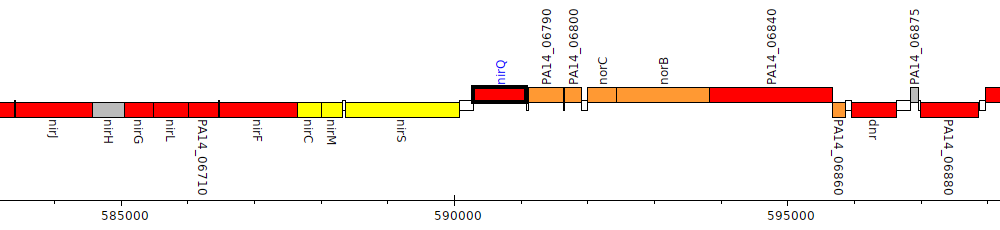
Pseudomonas aeruginosa UCBPP-PA14, PA14_06770 (nirQ)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07728
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF07728
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF08406 | CbbQ/NirQ/NorQ C-terminal | IPR013615 | CbbQ/NirQ/NorQ, C-terminal | 174 | 257 | 8.2E-30 |
| PANTHER | PTHR42759 | MOXR FAMILY PROTEIN | - | - | 13 | 214 | 1.1E-30 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 14 | 232 | 1.74E-33 |
| Pfam | PF07728 | AAA domain (dynein-related subfamily) | IPR011704 | ATPase, dynein-related, AAA domain | 27 | 161 | 1.8E-31 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 11 | 179 | 4.7E-30 |
| Coils | Coil | Coil | - | - | 185 | 205 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.