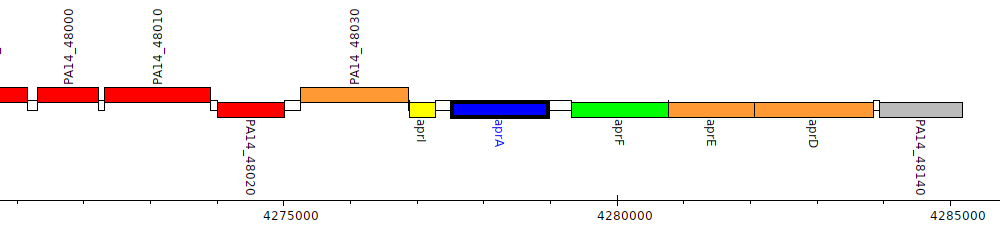
Pseudomonas aeruginosa UCBPP-PA14, PA14_48060 (aprA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF001205
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004222 | metalloendopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF001205
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:SM00235
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0005615 | extracellular space |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF001205
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008237 | metallopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SM00235
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008270 | zinc ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:SM00235
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pau01503 | Cationic antimicrobial peptide (CAMP) resistance | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00313 | NodO calcium binding signature | - | - | 369 | 377 | 3.9E-12 |
| Pfam | PF13583 | Metallo-peptidase family M12B Reprolysin-like | - | - | 170 | 230 | 1.8E-5 |
| FunFam | G3DSA:2.150.10.10:FF:000001 | Serralysin | - | - | 254 | 476 | 1.2E-117 |
| NCBIfam | NF035945 | NCBIFAM: serralysin family metalloprotease | - | - | 14 | 479 | 0.0 |
| Pfam | PF00353 | RTX calcium-binding nonapeptide repeat (4 copies) | IPR001343 | RTX calcium-binding nonapeptide repeat | 350 | 384 | 1.1E-9 |
| SUPERFAMILY | SSF55486 | Metalloproteases ("zincins"), catalytic domain | - | - | 14 | 255 | 7.58E-71 |
| SUPERFAMILY | SSF51120 | beta-Roll | IPR011049 | Serralysin-like metalloprotease, C-terminal | 256 | 478 | 4.32E-48 |
| Pfam | PF08548 | Peptidase M10 serralysin C terminal | IPR013858 | Peptidase M10 serralysin, C-terminal | 257 | 479 | 1.1E-81 |
| PRINTS | PR00313 | NodO calcium binding signature | - | - | 378 | 387 | 3.9E-12 |
| PIRSF | PIRSF001205 | Serralysin | IPR016294 | Peptidase M10B | 1 | 479 | 0.0 |
| CDD | cd04277 | ZnMc_serralysin_like | IPR034033 | Serralysin-like metallopeptidase domain | 65 | 256 | 1.61901E-45 |
| Gene3D | G3DSA:2.150.10.10 | - | IPR011049 | Serralysin-like metalloprotease, C-terminal | 21 | 476 | 0.0 |
| PRINTS | PR00313 | NodO calcium binding signature | - | - | 339 | 350 | 3.9E-12 |
| PRINTS | PR00313 | NodO calcium binding signature | - | - | 360 | 368 | 3.9E-12 |
| FunFam | G3DSA:3.40.390.10:FF:000046 | Serralysin | - | - | 27 | 259 | 5.8E-120 |
| Gene3D | G3DSA:3.40.390.10 | Collagenase (Catalytic Domain) | IPR024079 | Metallopeptidase, catalytic domain superfamily | 27 | 259 | 0.0 |
| SMART | SM00235 | col_5 | IPR006026 | Peptidase, metallopeptidase | 78 | 241 | 4.9E-29 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.