Pseudomonas aeruginosa PAO1, PA0317
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
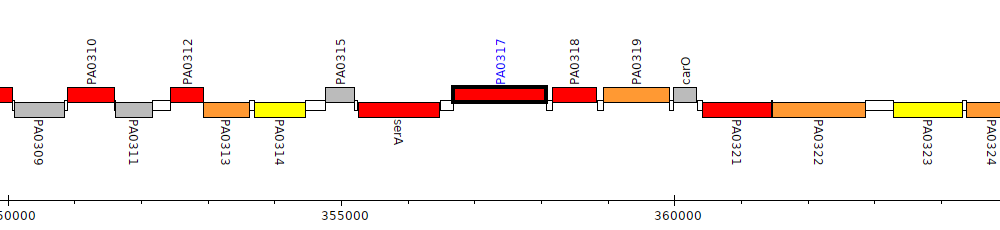
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0317
|
| Name |
|
| Replicon | chromosome |
| Genomic location | 356681 - 358075 (+ strand) |
| Transposon Mutants | 1 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
| Comment | PaD2HGDH is active with cobalt, manganese, nickel, or cadmium as an alternative metal |
| Comment | The FAD serves as the reaction center for hydride transfer |
| Comment | PA0317 is a D-2-hydroxyglutarate dehydrogenase that converts D-2-hydroxyglutarate to 2-ketoglutarate and its alternative substrate D-malate to oxaloacetate |
| Comment | Zinc and FAD-dependent enzyme whose electron acceptor is unknown and not oxygen. It uses 1 mole of zinc per mole of protein to orient and activate the substrate upon binding |
Cross-References
| RefSeq | NP_249008.1 |
| GI | 15595514 |
| Affymetrix | PA0317_at |
| Entrez | 878323 |
| GenBank | AAG03706.1 |
| INSDC | AAG03706.1 |
| NCBI Locus Tag | PA0317 |
| protein_id(GenBank) | gb|AAG03706.1|AE004469_11|gnl|PseudoCAP|PA0317 |
| TIGR | NTL03PA00318 |
| UniParc | UPI00000C5000 |
| UniProtKB Acc | Q9I6H4 |
| UniProtKB ID | Q9I6H4_PSEAE |
| UniRef100 | UniRef100_Q9I6H4 |
| UniRef50 | UniRef50_F5SNV8 |
| UniRef90 | UniRef90_Q9I6H4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
D-2-hydroxyglutarate dehydrogenase
|
| Synonyms |
D2HGDH |
| Evidence for Translation |
Detected by MALDI-TOF/TOF (PMID:19333994).
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -13.18 |
| Kyte-Doolittle Hydrophobicity Value | -0.079 |
| Molecular Weight (kDa) | 51.3 |
| Isoelectric Point (pI) | 4.88 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 549 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000314 (528 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0317
Search term: D-2-hydroxyglutarate dehydrogenase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
D-2-hydroxyglutarate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:28358]
E-value:
5.3e-72
Percent Identity:
34.4
|
References
|
Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen.
Imperi F, Ciccosanti F, Perdomo AB, Tiburzi F, Mancone C, Alonzi T, Ascenzi P, Piacentini M, Visca P, Fimia GM
Proteomics 2009 Apr;9(7):1901-15
PubMed ID: 19333994
|
|
Coupling between d-3-phosphoglycerate dehydrogenase and d-2-hydroxyglutarate dehydrogenase drives bacterial l-serine synthesis.
Zhang W, Zhang M, Gao C, Zhang Y, Ge Y, Guo S, Guo X, Zhou Z, Liu Q, Zhang Y, Ma C, Tao F, Xu P
Proc Natl Acad Sci U S A 2017 Sep 5;114(36):E7574-E7582
PubMed ID: 28827360
|
|
Kinetic and Bioinformatic Characterization of d-2-Hydroxyglutarate Dehydrogenase from Pseudomonas aeruginosa PAO1.
Quaye JA, Gadda G
Biochemistry 2020 Dec 29;59(51):4833-4844
PubMed ID: 33301690
|
|
Uncovering Zn(2+) as a cofactor of FAD-dependent Pseudomonas aeruginosa PAO1 d-2-hydroxyglutarate dehydrogenase.
Quaye JA, Gadda G
J Biol Chem 2023 Mar;299(3):103007
PubMed ID: 36775126
|
|
The Pseudomonas aeruginosa PAO1 metallo flavoprotein d-2-hydroxyglutarate dehydrogenase requires Zn(2+) for substrate orientation and activation.
Quaye JA, Gadda G
J Biol Chem 2023 Mar;299(3):103008
PubMed ID: 36775127
|