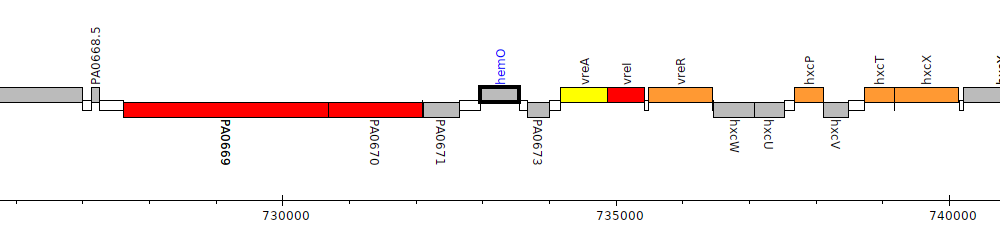
Pseudomonas aeruginosa PAO1, PA0672 (hemO)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0672
|
| Name |
hemO
Synonym: pigA |
| Replicon | chromosome |
| Genomic location | 732959 - 733555 (+ strand) |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_249363.1 |
| GI | 15595869 |
| Affymetrix | PA0672_at |
| Entrez | 880820 |
| GenBank | AAG04061.1 |
| INSDC | AAG04061.1 |
| NCBI Locus Tag | PA0672 |
| protein_id(GenBank) | gb|AAG04061.1|AE004502_4|gnl|PseudoCAP|PA0672 |
| TIGR | NTL03PA00673 |
| UniParc | UPI00000D4231 |
| UniProtKB Acc | G3XCZ8 |
| UniProtKB ID | G3XCZ8_PSEAE |
| UniRef100 | UniRef100_G3XCZ8 |
| UniRef50 | UniRef50_Q4K7S1 |
| UniRef90 | UniRef90_G3XCZ8 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
heme oxygenase
|
| Synonyms | |
| Evidence for Translation |
Verified using two-dimensional gel electrophoresis, MALDI-TOF MS and peptide mass mapping(pI:5.0, molecular weight:21.9, PMID:21674802)
|
| Charge (pH 7) | -8.03 |
| Kyte-Doolittle Hydrophobicity Value | -0.418 |
| Molecular Weight (kDa) | 22.0 |
| Isoelectric Point (pI) | 4.78 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 1SK7 | OXIDOREDUCTASE | 03/04/04 | Structural Basis for Novel Delta-Regioselective Heme Oxygenation in the Opportunistic Pathogen Pseudomonas aeruginosa | Pseudomonas aeruginosa | 1.6 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 23 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000651 (473 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0672
Search term: hemO
Search term: heme oxygenase
|
Human Homologs
References
|
Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase.
Zhu W, Wilks A, Stojiljkovic I
J Bacteriol 2000 Dec;182(23):6783-90
PubMed ID: 11073924
|
|
Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes.
Ochsner UA, Vasil ML
Proc Natl Acad Sci U S A 1996 Apr 30;93(9):4409-14
PubMed ID: 8633080
|
|
Proteomics of the oxidative stress response induced by hydrogen peroxide and paraquat reveals a novel AhpC-like protein in Pseudomonas aeruginosa.
Hare NJ, Scott NE, Shin EH, Connolly AM, Larsen MR, Palmisano G, Cordwell SJ
Proteomics 2011 Aug;11(15):3056-69
PubMed ID: 21674802
|
|
The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase.
Lansky IB, Lukat-Rodgers GS, Block D, Rodgers KR, Ratliff M, Wilks A
J Biol Chem 2006 May 12;281(19):13652-13662
PubMed ID: 16533806
|