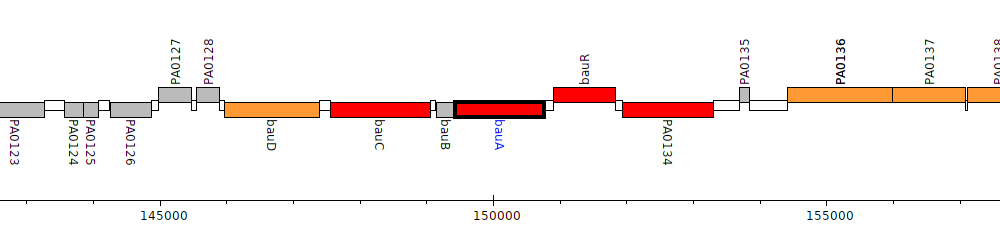
Pseudomonas aeruginosa PAO1, PA0132 (bauA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0132
|
| Name |
bauA
Synonym: oapT |
| Replicon | chromosome |
| Genomic location | 149425 - 150771 (- strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 5 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_248822.1 |
| GI | 15595330 |
| Affymetrix | PA0132_at |
| Entrez | 879350 |
| GenBank | AAG03522.1 |
| INSDC | AAG03522.1 |
| NCBI Locus Tag | PA0132 |
| protein_id(GenBank) | gb|AAG03522.1|AE004451_3|gnl|PseudoCAP|PA0132 |
| TIGR | NTL03PA00133 |
| UniParc | UPI00000C4F54 |
| UniProtKB Acc | Q9I700 |
| UniProtKB ID | BAUA_PSEAE |
| UniRef100 | UniRef100_Q9I700 |
| UniRef50 | UniRef50_Q9I700 |
| UniRef90 | UniRef90_Q9I700 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
Beta-alanine:pyruvate transaminase
|
| Synonyms |
omega-amino acid--pyruvate transaminase |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -3.78 |
| Kyte-Doolittle Hydrophobicity Value | -0.098 |
| Molecular Weight (kDa) | 48.4 |
| Isoelectric Point (pI) | 6.51 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 4B98 | TRANSFERASE | 09/03/12 | The structure of the omega aminotransferase from Pseudomonas aeruginosa | PSEUDOMONAS AERUGINOSA | 1.65 | X-RAY DIFFRACTION | 100.0 |
| 4B9B | TRANSFERASE | 09/03/12 | The structure of the omega aminotransferase from Pseudomonas aeruginosa | PSEUDOMONAS AERUGINOSA | 1.64 | X-RAY DIFFRACTION | 100.0 |
| 4BQ0 | TRANSFERASE | 05/29/13 | Pseudomonas aeruginosa beta-alanine:pyruvate aminotransferase holoenzyme without divalent cations on dimer-dimer interface | PSEUDOMONAS AERUGINOSA | 1.77 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 638 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000130 (850 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0132
Search term: bauA
Search term: Beta-alanine:pyruvate transaminase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
ornithine aminotransferase [Source:HGNC Symbol;Acc:HGNC:8091]
E-value:
6.5e-35
Percent Identity:
27.0
|
References
|
The primary structure of omega-amino acid:pyruvate aminotransferase.
Yonaha K, Nishie M, Aibara S
J Biol Chem 1992 Jun 25;267(18):12506-10
PubMed ID: 1618757
|
|
Functional characterization of seven γ-Glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and β-Alanine utilization in Pseudomonas aeruginosa PAO1.
Yao X, He W, Lu CD
J. Bacteriol. 2011 Aug;193(15):3923-30
PubMed ID: 21622750
|