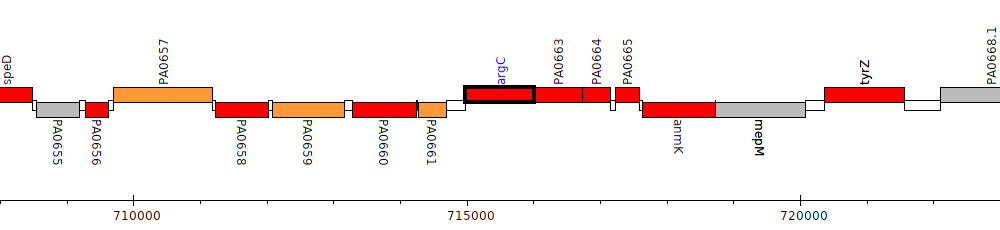
Pseudomonas aeruginosa PAO1, PA0662 (argC)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006520 | cellular amino acid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0008800 | beta-lactamase activity | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Molecular Function | GO:0051287 | NAD binding |
Inferred from Sequence Model
Term mapped from: InterPro:SM00859
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006526 | arginine biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00150
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003942 | N-acetyl-gamma-glutamyl-phosphate reductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00150
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
Inferred from Sequence Model
Term mapped from: InterPro:SM00859
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0008652 | cellular amino acid biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF02774
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0070401 | NADP+ binding |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00150
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0046983 | protein dimerization activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF02774
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00220 | Arginine biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | L-arginine biosynthesis IV (archaebacteria) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | pae01210 | 2-Oxocarboxylic acid metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | L-arginine biosynthesis III (via <i>N</i>-acetyl-L-citrulline) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | pae01230 | Biosynthesis of amino acids | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | GLUTORN-PWY | L-ornithine biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | ARGSYNBSUB-PWY | L-arginine biosynthesis II (acetyl cycle) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF51735 | NAD(P)-binding Rossmann-fold domains | IPR036291 | NAD(P)-binding domain superfamily | 2 | 172 | 1.74E-53 |
| Pfam | PF01118 | Semialdehyde dehydrogenase, NAD binding domain | IPR000534 | Semialdehyde dehydrogenase, NAD-binding | 3 | 142 | 3.2E-38 |
| Gene3D | G3DSA:3.30.360.10 | Dihydrodipicolinate Reductase; domain 2 | - | - | 148 | 312 | 4.9E-128 |
| Hamap | MF_00150 | N-acetyl-gamma-glutamyl-phosphate reductase [argC]. | IPR000706 | N-acetyl-gamma-glutamyl-phosphate reductase, type 1 | 2 | 338 | 43.662308 |
| SMART | SM00859 | Semialdhyde_dh_3 | IPR000534 | Semialdehyde dehydrogenase, NAD-binding | 3 | 142 | 8.1E-64 |
| Pfam | PF02774 | Semialdehyde dehydrogenase, dimerisation domain | IPR012280 | Semialdehyde dehydrogenase, dimerisation domain | 159 | 315 | 6.7E-17 |
| SUPERFAMILY | SSF55347 | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | - | - | 150 | 312 | 1.09E-51 |
| NCBIfam | TIGR01850 | JCVI: N-acetyl-gamma-glutamyl-phosphate reductase | IPR000706 | N-acetyl-gamma-glutamyl-phosphate reductase, type 1 | 2 | 344 | 0.0 |
| Gene3D | G3DSA:3.40.50.720 | - | - | - | 3 | 330 | 4.9E-128 |
| FunFam | G3DSA:3.30.360.10:FF:000014 | N-acetyl-gamma-glutamyl-phosphate reductase | - | - | 148 | 312 | 8.4E-52 |
| PANTHER | PTHR32338 | N-ACETYL-GAMMA-GLUTAMYL-PHOSPHATE REDUCTASE, CHLOROPLASTIC-RELATED-RELATED | - | - | 2 | 344 | 9.5E-126 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.