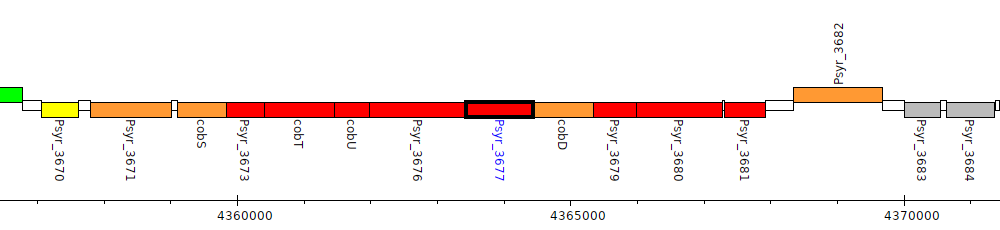
Pseudomonas syringae pv. syringae B728a, Psyr_3677
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0030170 | pyridoxal phosphate binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00155
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01140
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009236 | cobalamin biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01140
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009058 | biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00155
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | psb00860 | Porphyrin and chlorophyll metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | psb01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | aminopropanol phosphate biosynthesis I | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.640.10 | - | IPR015421 | Pyridoxal phosphate-dependent transferase, major domain | 42 | 237 | 4.5E-46 |
| Pfam | PF00155 | Aminotransferase class I and II | IPR004839 | Aminotransferase, class I/classII | 52 | 283 | 1.3E-26 |
| Gene3D | G3DSA:3.90.1150.10 | Aspartate Aminotransferase, domain 1 | IPR015422 | Pyridoxal phosphate-dependent transferase, small domain | 238 | 299 | 4.5E-46 |
| SUPERFAMILY | SSF53383 | PLP-dependent transferases | IPR015424 | Pyridoxal phosphate-dependent transferase | 3 | 326 | 3.06E-53 |
| NCBIfam | TIGR01140 | JCVI: threonine-phosphate decarboxylase CobD | IPR005860 | L-threonine-O-3-phosphate decarboxylase | 4 | 326 | 1.3E-104 |
| Coils | Coil | Coil | - | - | 241 | 261 | - |
| PANTHER | PTHR42885 | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE-RELATED | - | - | 15 | 265 | 4.8E-27 |
| CDD | cd00609 | AAT_like | - | - | 23 | 296 | 9.34826E-38 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.