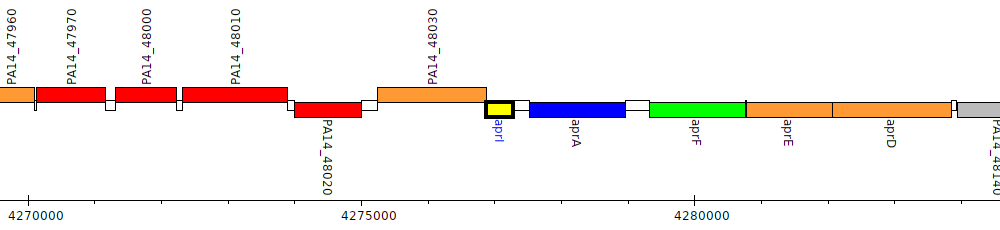
Pseudomonas aeruginosa UCBPP-PA14, PA14_48040 (aprI)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0010951 | negative regulation of endopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR01274
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008191 | metalloendopeptidase inhibitor activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR01274
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004866 | endopeptidase inhibitor activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF50882
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR01274 | Metalloprotease inhibitor signature | IPR022815 | Protease inhibitor | 107 | 120 | 5.9E-17 |
| PRINTS | PR01274 | Metalloprotease inhibitor signature | IPR022815 | Protease inhibitor | 68 | 79 | 5.9E-17 |
| SUPERFAMILY | SSF50882 | beta-Barrel protease inhibitors | IPR016085 | Protease inhibitor, beta-barrel domain | 26 | 128 | 3.14E-26 |
| PRINTS | PR01274 | Metalloprotease inhibitor signature | IPR022815 | Protease inhibitor | 26 | 42 | 5.9E-17 |
| Pfam | PF02974 | Protease inhibitor Inh | IPR021140 | Alkaline proteinase inhibitor/ Outer membrane lipoprotein Omp19 | 32 | 128 | 7.9E-23 |
| Gene3D | G3DSA:2.40.128.10 | - | - | - | 26 | 131 | 4.8E-32 |
| PRINTS | PR01274 | Metalloprotease inhibitor signature | IPR022815 | Protease inhibitor | 90 | 103 | 5.9E-17 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.