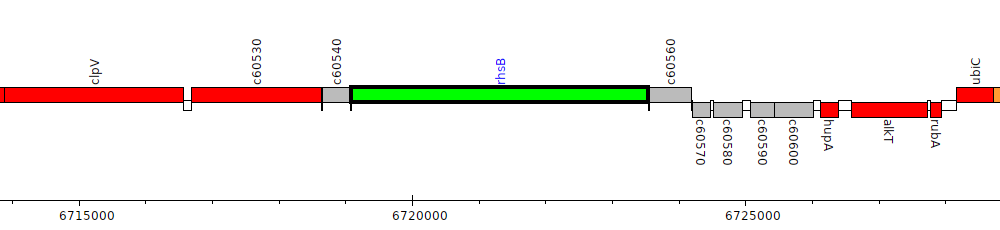
Pseudomonas protegens CHA0, PFLCHA0_c60550 (rhsB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR32305 | - | - | - | 894 | 1351 | 1.0E-37 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 614 | 644 | 1.8E-6 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 820 | 860 | 2.5E-6 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1295 | 1315 | 5.9E-13 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 694 | 726 | 0.001 |
| Gene3D | G3DSA:2.180.10.10 | - | - | - | 742 | 935 | 4.2E-31 |
| SUPERFAMILY | SSF63829 | Calcium-dependent phosphotriesterase | - | - | 594 | 846 | 1.31E-7 |
| Pfam | PF20148 | Domain of unknown function (DUF6531) | IPR045351 | Domain of unknown function DUF6531 | 304 | 383 | 5.9E-23 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 499 | 535 | 0.0066 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 588 | 625 | 3.2E-4 |
| Gene3D | G3DSA:2.180.10.10 | - | - | - | 480 | 741 | 4.2E-36 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 609 | 648 | 2.9E-7 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1318 | 1337 | 5.9E-13 |
| Gene3D | G3DSA:2.180.10.10 | - | - | - | 936 | 1349 | 1.3E-78 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 715 | 751 | 1.4E-6 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 736 | 773 | 2.0E-6 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 673 | 709 | 2.3E-8 |
| CDD | cd14742 | PAAR_RHS | - | - | 100 | 237 | 2.81631E-24 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 799 | 828 | 5.3E-5 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 1134 | 1168 | 0.008 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 820 | 856 | 8.0E-6 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 900 | 932 | 0.0035 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 736 | 776 | 1.7E-5 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 499 | 535 | 2.4E-6 |
| Gene3D | G3DSA:2.60.200.60 | - | - | - | 174 | 237 | 7.1E-12 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 122 | 152 | - |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 900 | 928 | 1.1E-4 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 781 | 818 | 0.0035 |
| Pfam | PF05488 | PAAR motif | IPR008727 | PAAR motif | 182 | 226 | 1.2E-7 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 658 | 692 | 1.6E-4 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 799 | 835 | 1.8E-7 |
| Pfam | PF03527 | RHS protein | IPR001826 | RHS protein | 1244 | 1280 | 3.4E-14 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1238 | 1258 | 5.9E-13 |
| NCBIfam | TIGR03696 | JCVI: RHS repeat-associated core domain | IPR022385 | Rhs repeat-associated core | 1271 | 1349 | 4.6E-27 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.