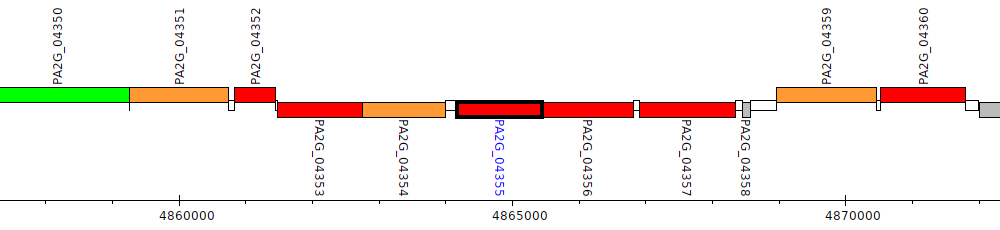
Pseudomonas aeruginosa 2192, PA2G_04355
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Cellular Component | GO:0005737 | cytoplasm |
Inferred from Sequence Model
Term mapped from: InterPro:PF01180
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors |
Inferred from Sequence Model
Term mapped from: InterPro:PF01180
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF54862 | 4Fe-4S ferredoxins | - | - | 318 | 402 | 3.52E-13 |
| Gene3D | G3DSA:3.20.20.70 | Aldolase class I | IPR013785 | Aldolase-type TIM barrel | 5 | 311 | 1.2E-95 |
| FunFam | G3DSA:3.20.20.70:FF:000027 | Dihydropyrimidine dehydrogenase [NADP(+)] | - | - | 5 | 312 | 5.6E-118 |
| Gene3D | G3DSA:3.30.70.20 | - | - | - | 317 | 415 | 1.8E-25 |
| Pfam | PF01180 | Dihydroorotate dehydrogenase | IPR005720 | Dihydroorotate dehydrogenase domain | 5 | 306 | 1.5E-27 |
| SUPERFAMILY | SSF51395 | FMN-linked oxidoreductases | - | - | 2 | 320 | 7.33E-76 |
| Pfam | PF14697 | 4Fe-4S dicluster domain | - | - | 339 | 401 | 4.5E-22 |
| PANTHER | PTHR43073 | DIHYDROPYRIMIDINE DEHYDROGENASE [NADP(+)] | - | - | 308 | 408 | 0.0 |
| NCBIfam | TIGR01037 | JCVI: dihydroorotate dehydrogenase family protein | IPR005720 | Dihydroorotate dehydrogenase domain | 5 | 320 | 2.6E-49 |
| CDD | cd02940 | DHPD_FMN | - | - | 4 | 302 | 0.0 |
| FunFam | G3DSA:3.30.70.20:FF:000087 | NAD-dependent dihydropyrimidine dehydrogenase subunit PreA | - | - | 309 | 412 | 7.3E-66 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.