Frequently Asked Questions
How should I cite the use of the Pseudomonas Genome Database?
Please cite our Nucleic Acids Research paper by Winsor et al. (2015) .
I am a researcher studying Pseudomonas. Where can I obtain the PAO1 strain that was sequenced, to use in my research? Where can I obtain the cosmids or other DNA constructs used in the genome sequencing project for Pseudomonas aeruginosa PAO1?
Requests for the P. aeruginosa PAO1 strain that was sequenced for the first Pseudomonas Genome Project should be sent to Dr. Stephen Lory at Harvard Medical School, Boston.
Requests for any other materials associated with the Pseudomonas Genome Project should be sent to Dr. Maynard Olson at the University of Washington, Seattle.
Where can I download a file containing the ORF DNA sequences for Pseudomonas strains?
You can download DNA sequences for genes in FASTA format by visiting our downloads page and clicking on the "Gene DNA" links.
How do I obtain the nucleotide sequence for an intergenic region?
There are a few ways you can go about doing this.
1) If you know the exact coordinates you are interested in, go to the sequence retrieval page and scroll down to a form that is titled "Retrieve a Specific Genomic Range from the Updated Database". Enter the start and stop coordinates of the region of interest and this will return all of the genomic DNA sequence within those coordinates.
2) Intergenic regions (both forward and reverse strand) can be downloaded in FASTA format by visiting the downloads page and clicking on the "Intergenic" link for individual strains.
3) Each gene card contains the nucleotide sequences for the upstream 500 bp and downstream 500 bp regions.
What is the difference between protein name confidence rating and localization confidence rating? What are their classifications based on?
The protein name confidence rating summarizes the degree of confidence in the function of a particular protein while the localization rating summarizes the degree of confidence in a protein’s subcellular localization.
The product name confidence rating can be summarized as follows:
Class 1: Function experimentally demonstrated in Pseudomonas aeruginosa.
Class 2: Function of highly similar gene experimentally demonstrated in another organism (and gene context consistent in terms of pathways it is involved in, if known).
Class 3: Function proposed based on presence of conserved amino acid motif, structural feature or limited sequence similarity to an experimentally studied gene.
Class 4: Homologs of previously reported genes of unknown function, or no similarity to any previously reported sequences.
The localization confidence rating can be summarized as follows:
Class 1: Subcellular localization experimentally demonstrated in Pseudomonas aeruginosa.
Class 2: Subcellular localization of highly similar gene experimentally demonstrated in another organism OR to a paralog experimentally demonstrated in the same organism. BLAST expect value of 10e-10 for query within 80-120% of subject length.
Class 3: Subcellular localization computationally predicted by PSORT.
What levels of homology are indicated by "strong homology" in level 2 for your annotation classification system?
There is no short answer to the question of what makes a gene Class 2. Homology counts, context counts, uniqueness of that gene in the genome counts, etc. On a genome annotation scale, consistency in how you decide counts, too. For the sake of space and focus, this issue was not fully addressed in the genome paper, however you may wish to contact those at Chiron (formerly of *PathoGenesis) involved in the initial genome annotation (see the genome paper) to speak with them about this further, as they took the lead role in the decision making for Class 2 verses Class 3 annotation designations.
Patient Questions about Pseudomonas infection
Please note that we are researchers involved in furthering the understanding of the biology of Pseudomonas aeruginosa. We are not qualified to answer questions regarding treatments for Cystic Fibrosis, and/or Pseudomonas infections. Any medically related questions should be referred to your personal primary care physician or other medical practitioner. For your benefit, we have provided answers to some frequently asked questions.I am taking a particular drug(s) to treat Pseudomonas infection and it's not working. Do you have any suggestions for a better treatment? Since we are research scientists and not physicians, we cannot provide you with any advice regarding treatment. Unfortunately, Pseudomonas infection can be difficult to treat in some instances; however we hope that our research will provide a better understanding of Pseudomonas aeruginosa so that new drugs or other therapies may be developed to prevent or cure infection. We recommend you bring up any concerns you have with your family doctor. Seek a second medical opinion if you feel strongly that you are being treated incorrectly.
*PathoGenesis Corporation, who played a major role in sequencing the first P. aeruginosa genome, is now a part of Chiron and continues its interest in the development of new anti-Pseudomonad drugs.
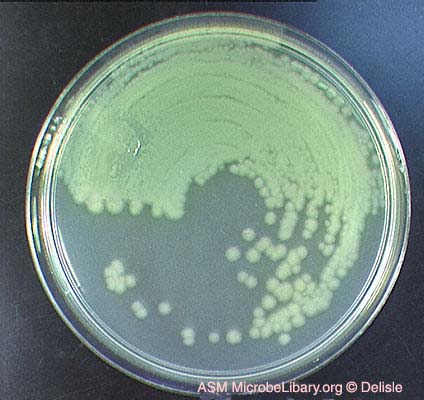
Pseudomonas aeruginosa and its green pigmentation
(Photo Credit: Gloria Delisle, Microbe Library, ASM).
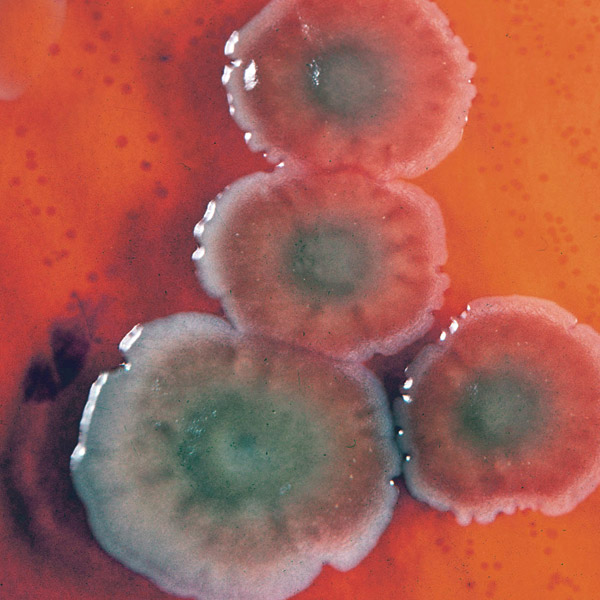
Pseudomonas aeruginosa
(Photo Credit: AJC ajcann.wordpress.com flickr site).