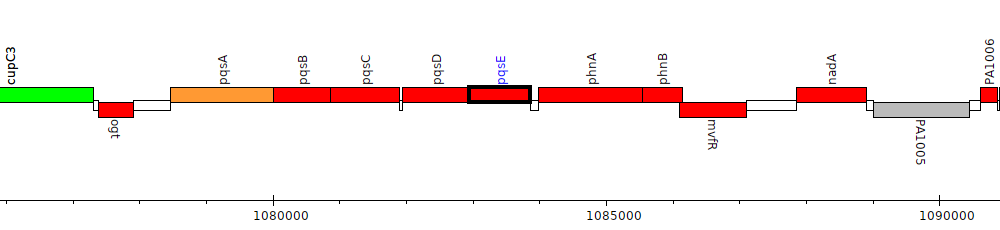
Pseudomonas aeruginosa PAO1, PA1000 (pqsE)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0051188 | obsolete cofactor biosynthetic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0044550 | secondary metabolite biosynthetic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
12426334 | Reviewed by curator |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Coils | Coil | Coil | - | - | 102 | 122 | - |
| CDD | cd07725 | TTHA1429-like_MBL-fold | - | - | 21 | 231 | 1.44485E-52 |
| PANTHER | PTHR42951 | METALLO-BETA-LACTAMASE DOMAIN-CONTAINING | - | - | 20 | 229 | 3.8E-25 |
| FunFam | G3DSA:3.60.15.10:FF:000088 | Quinolone signal response protein | - | - | 1 | 301 | 0.0 |
| Gene3D | G3DSA:3.60.15.10 | - | IPR036866 | Ribonuclease Z/Hydroxyacylglutathione hydrolase-like | 1 | 301 | 5.3E-92 |
| SMART | SM00849 | Lactamase_B_5a | IPR001279 | Metallo-beta-lactamase | 21 | 221 | 2.0E-26 |
| SUPERFAMILY | SSF56281 | Metallo-hydrolase/oxidoreductase | IPR036866 | Ribonuclease Z/Hydroxyacylglutathione hydrolase-like | 14 | 269 | 7.6E-53 |
| Pfam | PF00753 | Metallo-beta-lactamase superfamily | IPR001279 | Metallo-beta-lactamase | 18 | 221 | 5.2E-27 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.