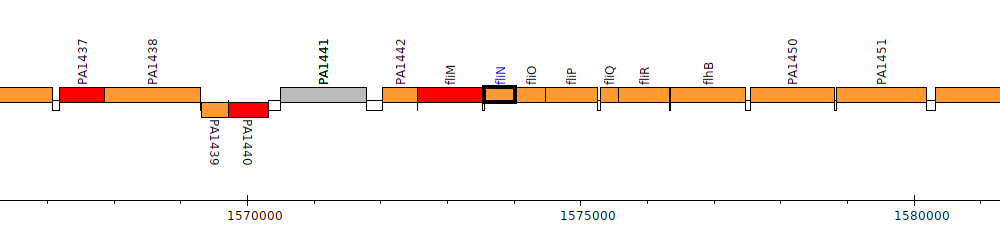
Pseudomonas aeruginosa PAO1, PA1444 (fliN)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0050896 | response to stimulus | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0006935 | chemotaxis | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0006928 | movement of cell or subcellular component | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0071973 | bacterial-type flagellum-dependent cell motility |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02480
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0009425 | bacterial-type flagellum basal body |
Inferred from Sequence Model
Term mapped from: InterPro:PR00956
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006935 | chemotaxis |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02480
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003774 | motor activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00956
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02480
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0009288 | bacterial-type flagellum |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02480
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Flagella assembly |
ECO:0000037
not_recorded |
|||
| KEGG | pae02040 | Flagellar assembly | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae02030 | Bacterial chemotaxis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:2.30.330.10 | - | IPR036429 | SpoA-like superfamily | 70 | 157 | 6.4E-31 |
| PANTHER | PTHR43484 | - | - | - | 37 | 157 | 1.1E-35 |
| PRINTS | PR00956 | Flagellar motor switch protein FliN signature | IPR001172 | Flagellar motor switch FliN/Type III secretion HrcQb | 94 | 110 | 2.6E-28 |
| PRINTS | PR00956 | Flagellar motor switch protein FliN signature | IPR001172 | Flagellar motor switch FliN/Type III secretion HrcQb | 110 | 123 | 2.6E-28 |
| Pfam | PF01052 | Type III flagellar switch regulator (C-ring) FliN C-term | IPR001543 | Flagellar motor switch protein FliN-like, C-terminal domain | 78 | 147 | 4.3E-25 |
| PRINTS | PR00956 | Flagellar motor switch protein FliN signature | IPR001172 | Flagellar motor switch FliN/Type III secretion HrcQb | 123 | 138 | 2.6E-28 |
| NCBIfam | TIGR02480 | JCVI: flagellar motor switch protein FliN | IPR012826 | Flagellar motor switch FliN | 74 | 149 | 1.1E-32 |
| FunFam | G3DSA:2.30.330.10:FF:000001 | Flagellar motor switch protein FliN | - | - | 66 | 157 | 8.3E-50 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 24 | 67 | - |
| PRINTS | PR00956 | Flagellar motor switch protein FliN signature | IPR001172 | Flagellar motor switch FliN/Type III secretion HrcQb | 77 | 93 | 2.6E-28 |
| SUPERFAMILY | SSF101801 | Surface presentation of antigens (SPOA) | IPR036429 | SpoA-like superfamily | 74 | 155 | 1.2E-25 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.