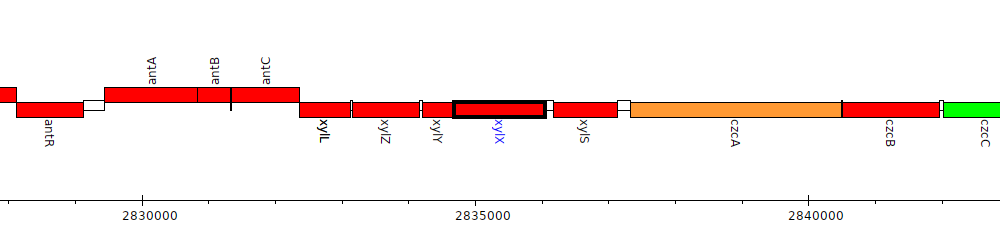
Pseudomonas aeruginosa PAO1, PA2518 (xylX)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0015976 | carbon utilization | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0044237 | cellular metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00848
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005506 | iron ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00848
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0051537 | 2 iron, 2 sulfur cluster binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00848
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Aromatic compound catabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae01220 | Degradation of aromatic compounds | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00362 | Benzoate degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00622 | Xylene degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00364 | Fluorobenzoate degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 189 | 211 | 6.1E-46 |
| Pfam | PF00355 | Rieske [2Fe-2S] domain | IPR017941 | Rieske [2Fe-2S] iron-sulphur domain | 45 | 129 | 1.3E-20 |
| Gene3D | G3DSA:2.102.10.10 | - | IPR036922 | Rieske [2Fe-2S] iron-sulphur domain superfamily | 45 | 172 | 9.4E-102 |
| CDD | cd08879 | RHO_alpha_C_AntDO-like | - | - | 194 | 434 | 4.93461E-87 |
| PANTHER | PTHR43756 | CHOLINE MONOOXYGENASE, CHLOROPLASTIC | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 6 | 442 | 9.8E-71 |
| NCBIfam | TIGR03229 | JCVI: benzoate 1,2-dioxygenase large subunit | IPR017639 | Benzoate 1,2-dioxygenase, large subunit | 9 | 438 | 0.0 |
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 60 | 75 | 6.1E-46 |
| CDD | cd03542 | Rieske_RO_Alpha_HBDO | - | - | 46 | 168 | 1.34621E-90 |
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 102 | 122 | 6.1E-46 |
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 30 | 55 | 6.1E-46 |
| SUPERFAMILY | SSF55961 | Bet v1-like | - | - | 164 | 445 | 1.33E-70 |
| Pfam | PF00848 | Ring hydroxylating alpha subunit (catalytic domain) | IPR015879 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit, C-terminal domain | 196 | 385 | 2.7E-18 |
| SUPERFAMILY | SSF50022 | ISP domain | IPR036922 | Rieske [2Fe-2S] iron-sulphur domain superfamily | 11 | 164 | 9.56E-45 |
| Gene3D | G3DSA:3.90.380.10 | - | - | - | 23 | 395 | 9.4E-102 |
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 81 | 101 | 6.1E-46 |
| PRINTS | PR00090 | Ring hydroxylating dioxygenase alpha-subunit signature | IPR001663 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit | 148 | 174 | 6.1E-46 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.