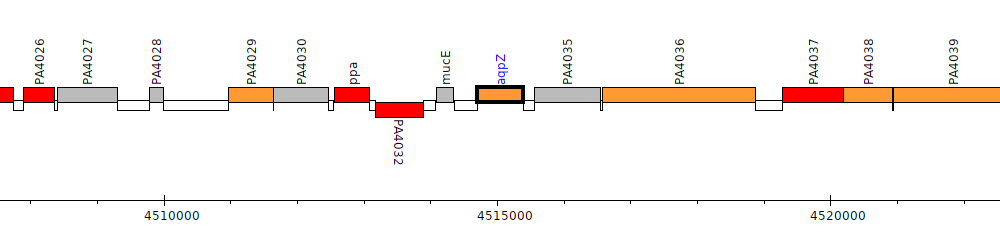
Pseudomonas aeruginosa PAO1, PA4034 (aqpZ)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Cellular Component | GO:0016020 | membrane | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
7493926 | Reviewed by curator |
| Biological Process | GO:0006810 | transport | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0015250 | water channel activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
7493926 | Reviewed by curator |
| Biological Process | GO:0006833 | water transport |
Inferred from Sequence Model
Term mapped from: InterPro:MF_01146
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PF00230
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0015267 | channel activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00230
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055085 | transmembrane transport |
Inferred from Sequence Model
Term mapped from: InterPro:PF00230
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0015250 | water channel activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_01146
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 204 | 224 | 1.2E-50 |
| Gene3D | G3DSA:1.20.1080.10 | Glycerol uptake facilitator protein. | IPR023271 | Aquaporin-like | 1 | 226 | 7.4E-75 |
| NCBIfam | TIGR00861 | JCVI: MIP family channel protein | IPR000425 | Major intrinsic protein | 6 | 221 | 7.2E-57 |
| FunFam | G3DSA:1.20.1080.10:FF:000007 | Aquaporin Z | - | - | 1 | 229 | 1.2E-118 |
| CDD | cd00333 | MIP | IPR000425 | Major intrinsic protein | 2 | 224 | 3.42773E-62 |
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 42 | 66 | 1.2E-50 |
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 79 | 98 | 1.2E-50 |
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 165 | 187 | 1.2E-50 |
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 2 | 21 | 1.2E-50 |
| PANTHER | PTHR19139 | AQUAPORIN TRANSPORTER | IPR034294 | Aquaporin transporter | 3 | 225 | 8.8E-53 |
| PRINTS | PR00783 | Major intrinsic protein family signature | IPR000425 | Major intrinsic protein | 135 | 153 | 1.2E-50 |
| Hamap | MF_01146 | Aquaporin Z [aqpZ]. | IPR023743 | Aquaporin Z | 1 | 228 | 35.102604 |
| Pfam | PF00230 | Major intrinsic protein | IPR000425 | Major intrinsic protein | 2 | 221 | 5.0E-46 |
| SUPERFAMILY | SSF81338 | Aquaporin-like | IPR023271 | Aquaporin-like | 1 | 225 | 6.54E-66 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.