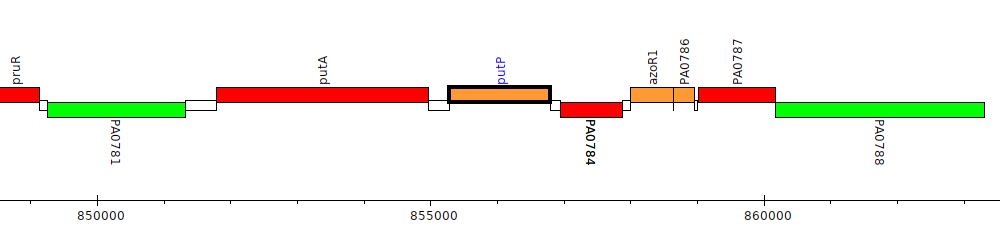
Pseudomonas aeruginosa PAO1, PA0783 (putP)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0015193 | L-proline transmembrane transporter activity | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
12270821 | Reviewed by curator |
| Biological Process | GO:0010133 | proline catabolic process to glutamate | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
12270821 | Reviewed by curator |
| Biological Process | GO:1904271 | L-proline import across plasma membrane | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
18833300 | Reviewed by curator |
| Molecular Function | GO:0031402 | sodium ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd11475
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0022857 | transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00474
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006814 | sodium ion transport |
Inferred from Sequence Model
Term mapped from: InterPro:cd11475
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005298 | proline:sodium symporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd11475
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:cd11475
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0015824 | proline transport |
Inferred from Sequence Model
Term mapped from: InterPro:cd11475
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055085 | transmembrane transport |
Inferred from Sequence Model
Term mapped from: InterPro:PF00474
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR00813 | JCVI: sodium/solute symporter | IPR001734 | Sodium/solute symporter | 35 | 437 | 2.7E-123 |
| NCBIfam | TIGR02121 | JCVI: sodium/proline symporter PutP | IPR011851 | Sodium/proline symporter | 6 | 491 | 0.0 |
| CDD | cd11475 | SLC5sbd_PutP | IPR011851 | Sodium/proline symporter | 8 | 473 | 0.0 |
| PANTHER | PTHR48086 | SODIUM/PROLINE SYMPORTER-RELATED | - | - | 2 | 491 | 7.2E-126 |
| Gene3D | G3DSA:1.20.1730.10 | Sodium/glucose cotransporter | IPR038377 | Sodium/glucose symporter superfamily | 24 | 501 | 1.7E-106 |
| FunFam | G3DSA:1.20.1730.10:FF:000002 | Sodium/proline symporter | - | - | 22 | 495 | 0.0 |
| Pfam | PF00474 | Sodium:solute symporter family | IPR001734 | Sodium/solute symporter | 35 | 437 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.