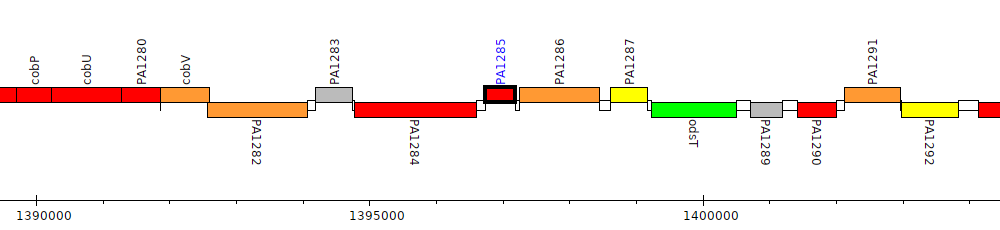
Pseudomonas aeruginosa PAO1, PA1285
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00598
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:PR00598
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF46785 | Winged helix DNA-binding domain | IPR036390 | Winged helix DNA-binding domain superfamily | 13 | 146 | 4.18E-30 |
| Pfam | PF12802 | MarR family | IPR000835 | MarR-type HTH domain | 42 | 98 | 1.9E-10 |
| SMART | SM00347 | marrlong4 | IPR000835 | MarR-type HTH domain | 36 | 135 | 1.4E-11 |
| PRINTS | PR00598 | Bacterial regulatory protein MarR family signature | IPR000835 | MarR-type HTH domain | 125 | 145 | 1.3E-5 |
| Gene3D | G3DSA:1.10.10.10 | - | IPR036388 | Winged helix-like DNA-binding domain superfamily | 14 | 148 | 9.7E-34 |
| PANTHER | PTHR33164 | TRANSCRIPTIONAL REGULATOR, MARR FAMILY | IPR039422 | Transcription regulators MarR/SlyA-like | 16 | 147 | 3.3E-20 |
| PRINTS | PR00598 | Bacterial regulatory protein MarR family signature | IPR000835 | MarR-type HTH domain | 95 | 111 | 1.3E-5 |
| PRINTS | PR00598 | Bacterial regulatory protein MarR family signature | IPR000835 | MarR-type HTH domain | 60 | 76 | 1.3E-5 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.