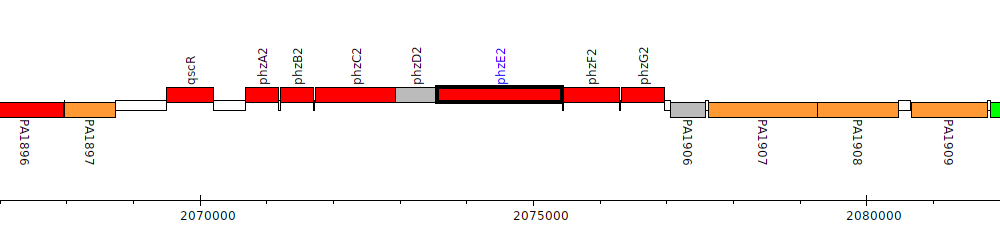
Pseudomonas aeruginosa PAO1, PA1903 (phzE2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0002047 | phenazine biosynthetic process | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
11591691 | Reviewed by curator |
| Biological Process | GO:0009058 | biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR11236
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Phenazine biosynthesis |
ECO:0000037
not_recorded |
|||
| PseudoCyc | PWYB-5652 | Phenazine biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF52317 | Class I glutamine amidotransferase-like | IPR029062 | Class I glutamine amidotransferase-like | 437 | 624 | 1.12E-22 |
| PRINTS | PR00096 | Glutamine amidotransferase superfamily signature | - | - | 478 | 487 | 2.8E-9 |
| Gene3D | G3DSA:3.60.120.10 | Anthranilate synthase | IPR005801 | ADC synthase | 4 | 391 | 1.5E-94 |
| SUPERFAMILY | SSF56322 | ADC synthase | IPR005801 | ADC synthase | 66 | 390 | 1.07E-66 |
| PRINTS | PR00097 | Anthranilate synthase component II signature | - | - | 597 | 610 | 3.6E-8 |
| Gene3D | G3DSA:3.40.50.880 | - | IPR029062 | Class I glutamine amidotransferase-like | 436 | 626 | 5.4E-45 |
| Pfam | PF00117 | Glutamine amidotransferase class-I | IPR017926 | Glutamine amidotransferase | 439 | 618 | 2.9E-25 |
| PRINTS | PR00097 | Anthranilate synthase component II signature | - | - | 437 | 451 | 3.6E-8 |
| PRINTS | PR00096 | Glutamine amidotransferase superfamily signature | - | - | 511 | 522 | 2.8E-9 |
| PRINTS | PR00099 | Carbamoyl-phosphate synthase protein GATase domain signature | - | - | 511 | 527 | 5.7E-5 |
| PRINTS | PR00096 | Glutamine amidotransferase superfamily signature | - | - | 597 | 610 | 2.8E-9 |
| PRINTS | PR00097 | Anthranilate synthase component II signature | - | - | 511 | 522 | 3.6E-8 |
| Pfam | PF00425 | chorismate binding enzyme | IPR015890 | Chorismate-utilising enzyme, C-terminal | 122 | 381 | 4.6E-49 |
| CDD | cd01743 | GATase1_Anthranilate_Synthase | IPR006221 | Anthranilate synthase/para-aminobenzoate synthase like domain | 437 | 618 | 1.23634E-49 |
| PRINTS | PR00099 | Carbamoyl-phosphate synthase protein GATase domain signature | - | - | 475 | 489 | 5.7E-5 |
| PANTHER | PTHR11236 | AMINOBENZOATE/ANTHRANILATE SYNTHASE | IPR019999 | Anthranilate synthase component I-like | 116 | 394 | 2.0E-52 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.