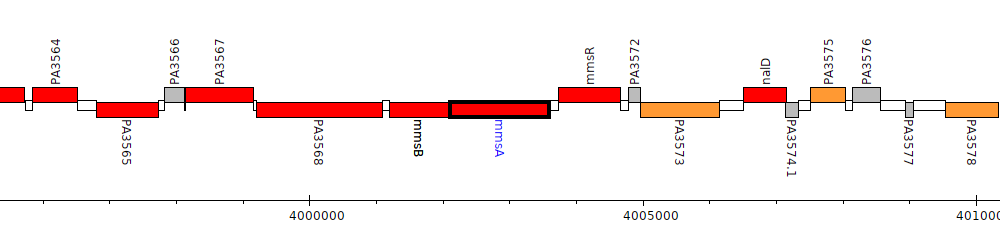
Pseudomonas aeruginosa PAO1, PA3570 (mmsA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006520 | cellular amino acid metabolic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0015976 | carbon utilization | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0006552 | leucine catabolic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0019541 | propionate metabolic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Molecular Function | GO:0004040 | amidase activity | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0006550 | isoleucine catabolic process | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00171
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004491 | methylmalonate-semialdehyde dehydrogenase (acylating) activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd07085
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.40.309.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Propanoate metabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae00410 | beta-Alanine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00562 | Inositol phosphate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00640 | Propanoate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | VALDEG-PWY | L-valine degradation I | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00280 | Valine, leucine and isoleucine degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Valine, leucine and isoleucine degradation |
ECO:0000037
not_recorded |
|||
| KEGG | pae01200 | Carbon metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.309.10 | Aldehyde Dehydrogenase; Chain A, domain 2 | IPR016163 | Aldehyde dehydrogenase, C-terminal | 250 | 450 | 0.0 |
| NCBIfam | TIGR01722 | JCVI: CoA-acylating methylmalonate-semialdehyde dehydrogenase | IPR010061 | Methylmalonate-semialdehyde dehydrogenase | 5 | 481 | 0.0 |
| Pfam | PF00171 | Aldehyde dehydrogenase family | IPR015590 | Aldehyde dehydrogenase domain | 18 | 477 | 0.0 |
| Coils | Coil | Coil | - | - | 41 | 61 | - |
| PANTHER | PTHR43866 | MALONATE-SEMIALDEHYDE DEHYDROGENASE | IPR010061 | Methylmalonate-semialdehyde dehydrogenase | 4 | 494 | 0.0 |
| Gene3D | G3DSA:3.40.605.10 | Aldehyde Dehydrogenase; Chain A, domain 1 | IPR016162 | Aldehyde dehydrogenase, N-terminal | 25 | 458 | 0.0 |
| FunFam | G3DSA:3.40.309.10:FF:000002 | Methylmalonate-semialdehyde dehydrogenase (Acylating) | - | - | 250 | 450 | 2.3E-89 |
| CDD | cd07085 | ALDH_F6_MMSDH | IPR010061 | Methylmalonate-semialdehyde dehydrogenase | 5 | 481 | 0.0 |
| FunFam | G3DSA:3.40.605.10:FF:000003 | Methylmalonate-semialdehyde dehydrogenase [acylating] | - | - | 9 | 256 | 1.9E-108 |
| SUPERFAMILY | SSF53720 | ALDH-like | IPR016161 | Aldehyde/histidinol dehydrogenase | 3 | 483 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.