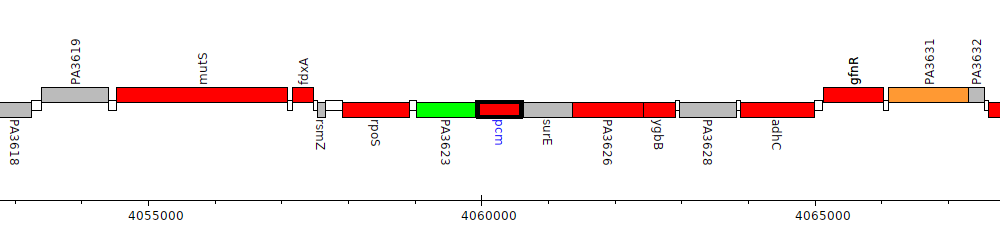
Pseudomonas aeruginosa PAO1, PA3624 (pcm)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3624
|
| Name |
pcm
|
| Replicon | chromosome |
| Genomic location | 4059957 - 4060592 (- strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252314.1 |
| GI | 15598820 |
| Affymetrix | PA3624_pcm_at |
| DNASU | PaCD00007586 |
| Entrez | 880441 |
| GenBank | AAG07012.1 |
| INSDC | BAA05129.1 |
| NCBI Locus Tag | PA3624 |
| protein_id(GenBank) | gb|AAG07012.1|AE004782_10|gnl|PseudoCAP|PA3624 |
| TIGR | NTL03PA03624 |
| UniParc | UPI0000131AE7 |
| UniProtKB Acc | P45683 |
| UniProtKB ID | PIMT_PSEAE |
| UniRef100 | UniRef100_B7V8C3 |
| UniRef50 | UniRef50_A6V1G4 |
| UniRef90 | UniRef90_A6V1G4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
L-isoaspartate protein carboxylmethyltransferase type II
|
| Synonyms | |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -2.04 |
| Kyte-Doolittle Hydrophobicity Value | -0.027 |
| Molecular Weight (kDa) | 23.4 |
| Isoelectric Point (pI) | 6.13 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 418 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002050 (534 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3624
Search term: pcm
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase [Source:HGNC Symbol;Acc:HGNC:8728]
E-value:
5.8e-18
Percent Identity:
36.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase [Source:HGNC Symbol;Acc:HGNC:8728]
E-value:
5.8e-18
Percent Identity:
36.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase [Source:HGNC Symbol;Acc:HGNC:8728]
E-value:
5.8e-18
Percent Identity:
36.0
|
References
|
Purification, gene cloning, and sequence analysis of an L-isoaspartyl protein carboxyl methyltransferase from Escherichia coli.
Fu JC, Ding L, Clarke S
J Biol Chem 1991 Aug 5;266(22):14562-72
PubMed ID: 1860862
|
|
The L-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses.
Visick JE, Cai H, Clarke S
J Bacteriol 1998 May;180(10):2623-9
PubMed ID: 9573145
|
|
Isoaspartate in ribosomal protein S11 of Escherichia coli.
David CL, Keener J, Aswad DW
J Bacteriol 1999 May;181(9):2872-7
PubMed ID: 10217780
|