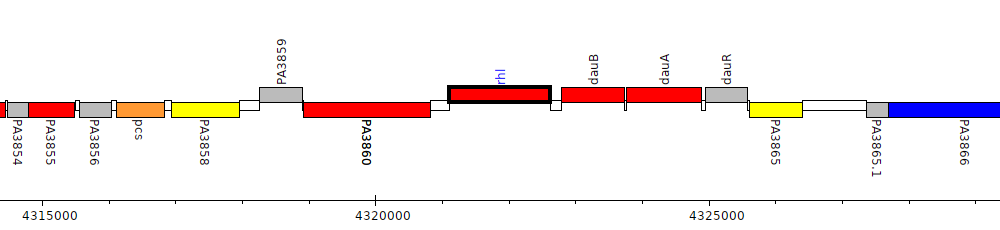
Pseudomonas aeruginosa PAO1, PA3861 (rhl)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0016070 | RNA metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00270
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003724 | RNA helicase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00661
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003676 | nucleic acid binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00270
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae03018 | RNA degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 88 | 333 | 2.0E-74 |
| SMART | SM00490 | helicmild6 | IPR001650 | Helicase, C-terminal | 381 | 462 | 2.0E-32 |
| CDD | cd00268 | DEADc | - | - | 130 | 331 | 1.49651E-92 |
| SMART | SM00487 | ultradead3 | IPR014001 | Helicase superfamily 1/2, ATP-binding domain | 138 | 345 | 4.8E-52 |
| Hamap | MF_00661 | ATP-dependent RNA helicase RhlB [rhlB]. | IPR023554 | ATP-dependent RNA helicase RhlB type | 118 | 500 | 53.172462 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 336 | 502 | 6.0E-48 |
| CDD | cd18787 | SF2_C_DEAD | - | - | 342 | 470 | 2.02791E-58 |
| Pfam | PF00270 | DEAD/DEAH box helicase | IPR011545 | DEAD/DEAH box helicase domain | 143 | 318 | 2.9E-48 |
| PANTHER | PTHR47959 | ATP-DEPENDENT RNA HELICASE RHLE-RELATED | - | - | 116 | 502 | 4.3E-124 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 40 | 102 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 1 | 102 | - |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 194 | 478 | 1.84E-72 |
| Pfam | PF00271 | Helicase conserved C-terminal domain | IPR001650 | Helicase, C-terminal | 353 | 462 | 7.5E-30 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.