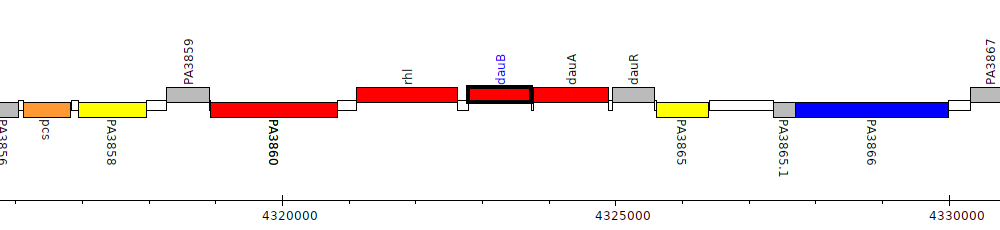
Pseudomonas aeruginosa PAO1, PA3862 (dauB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Operons
|
Operon name:
dauBAR operon
(3
gene members)
Experiment
Attributes
method:
purification of transcriptional regulutors, DauR and ArgR. Gel retardation assasys with the regulatory region.
Evidence
Transcription unit experimentally validated in a strain of the same species. The start coordinate will often be based on an experimentally validated transcription start site. For protein-coding genes, unless otherwise specified, the stop will be based on the stop codon.
References
Regulation of the dauBAR operon and characterization of D-amino acid dehydrogenase DauA in arginine and lysine catabolism of Pseudomonas aeruginosa PAO1.
Li C, Yao X, Lu CD
Microbiology (Reading, Engl.) 2010 Jan;156(Pt 1):60-71
PubMed ID: 19850617
Cross-References
PseudoCAP:
41781
|
||||||||||||||||||||||||||||||||
|
Operon name:
dauB-dauA-dauR
(3
gene members)
Evidence
Computationally-predicted.
References
DOOR: a database for prokaryotic operons.
Mao F, Dam P, Chou J, Olman V, Xu Y
Nucleic Acids Res. 2009 Jan;37(Database issue):D459-63
PubMed ID: 18988623
Cross-References
DOOR:
13891
|