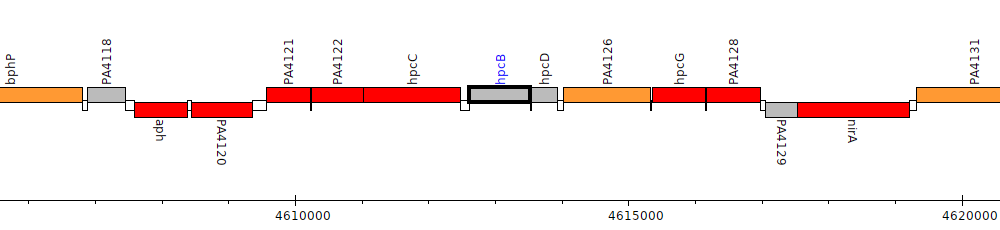
Pseudomonas aeruginosa PAO1, PA4124 (hpcB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0015976 | carbon utilization | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0008198 | ferrous iron binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF02900
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF02900
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006725 | cellular aromatic compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF02900
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008687 | 3,4-dihydroxyphenylacetate 2,3-dioxygenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd07370
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Aromatic compound catabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | 3-HYDROXYPHENYLACETATE-DEGRADATION-PWY | 4-hydroxyphenylacetate degradation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00350 | Tyrosine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01220 | Degradation of aromatic compounds | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR02298 | JCVI: 3,4-dihydroxyphenylacetate 2,3-dioxygenase | IPR011984 | 3,4-dihydroxyphenylacetate 2,3-dioxygenase | 1 | 282 | 0.0 |
| CDD | cd07370 | HPCD | IPR011984 | 3,4-dihydroxyphenylacetate 2,3-dioxygenase | 3 | 282 | 0.0 |
| PANTHER | PTHR30096 | UNCHARACTERIZED | - | - | 14 | 280 | 4.7E-22 |
| Gene3D | G3DSA:3.40.830.10 | - | - | - | 3 | 281 | 3.2E-61 |
| Pfam | PF02900 | Catalytic LigB subunit of aromatic ring-opening dioxygenase | IPR004183 | Extradiol ring-cleavage dioxygenase, class III enzyme, subunit B | 7 | 280 | 1.0E-79 |
| SUPERFAMILY | SSF53213 | LigB-like | - | - | 12 | 280 | 5.1E-50 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.