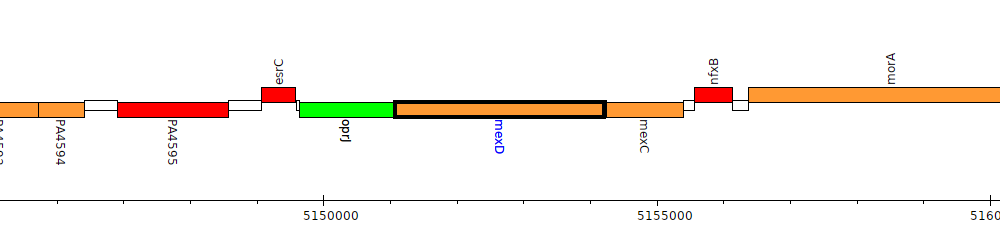
Pseudomonas aeruginosa PAO1, PA4598 (mexD)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0042493 | response to drug | Inferred from Experiment | ECO:0000269 experimental evidence used in manual assertion |
12654738 | Reviewed by curator |
| Cellular Component | GO:0016020 | membrane | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Biological Process | GO:0006810 | transport | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Molecular Function | GO:0042910 | xenobiotic transmembrane transporter activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
8878035 | Reviewed by curator |
| Biological Process | GO:0006811 | ion transport | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
||
| Biological Process | GO:0046677 | response to antibiotic | Inferred from Direct Assay | ECO:0000002 direct assay evidence |
||
| Molecular Function | GO:0015562 | efflux transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00915
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PF00873
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0022857 | transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00873
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0042908 | xenobiotic transport |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00915
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055085 | transmembrane transport |
Inferred from Sequence Model
Term mapped from: InterPro:PF00873
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR32063 | - | IPR001036 | Acriflavin resistance protein | 2 | 1027 | 0.0 |
| Gene3D | G3DSA:1.20.1640.10 | Multidrug efflux transporter AcrB transmembrane domain | - | - | 7 | 517 | 0.0 |
| SUPERFAMILY | SSF82693 | Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains | - | - | 148 | 332 | 4.24E-21 |
| SUPERFAMILY | SSF82866 | Multidrug efflux transporter AcrB transmembrane domain | - | - | 298 | 499 | 1.05E-49 |
| SUPERFAMILY | SSF82714 | Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains | IPR027463 | Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains | 721 | 806 | 5.36E-20 |
| FunFam | G3DSA:1.20.1640.10:FF:000001 | Efflux pump membrane transporter | - | - | 323 | 512 | 4.9E-71 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 554 | 571 | 9.0E-100 |
| Gene3D | G3DSA:1.20.1640.10 | Multidrug efflux transporter AcrB transmembrane domain | - | - | 518 | 1021 | 0.0 |
| NCBIfam | TIGR00915 | JCVI: efflux RND transporter permease subunit | IPR004764 | Multidrug resistance protein MdtF-like | 1 | 1038 | 0.0 |
| FunFam | G3DSA:3.30.70.1430:FF:000001 | Efflux pump membrane transporter | - | - | 38 | 135 | 1.2E-28 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 365 | 386 | 9.0E-100 |
| Gene3D | G3DSA:3.30.2090.10 | Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains | IPR027463 | Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains | 182 | 280 | 0.0 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 621 | 635 | 9.0E-100 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 393 | 417 | 9.0E-100 |
| Gene3D | G3DSA:3.30.70.1440 | Multidrug efflux transporter AcrB pore domain | - | - | 671 | 869 | 0.0 |
| Pfam | PF00873 | AcrB/AcrD/AcrF family | IPR001036 | Acriflavin resistance protein | 1 | 1023 | 0.0 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 447 | 470 | 9.0E-100 |
| Gene3D | G3DSA:3.30.70.1430 | Multidrug efflux transporter AcrB pore domain | - | - | 38 | 819 | 0.0 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 338 | 361 | 9.0E-100 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 36 | 54 | 9.0E-100 |
| Gene3D | G3DSA:3.30.2090.10 | Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains | IPR027463 | Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains | 723 | 811 | 0.0 |
| SUPERFAMILY | SSF82714 | Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains | IPR027463 | Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains | 185 | 274 | 3.53E-20 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 472 | 495 | 9.0E-100 |
| SUPERFAMILY | SSF82693 | Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains | - | - | 567 | 670 | 4.03E-24 |
| SUPERFAMILY | SSF82693 | Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains | - | - | 38 | 129 | 2.09E-23 |
| Gene3D | G3DSA:3.30.70.1430 | Multidrug efflux transporter AcrB pore domain | - | - | 572 | 670 | 0.0 |
| SUPERFAMILY | SSF82866 | Multidrug efflux transporter AcrB transmembrane domain | - | - | 804 | 1026 | 6.28E-47 |
| Gene3D | G3DSA:3.30.70.1320 | Multidrug efflux transporter AcrB pore domain like | - | - | 133 | 333 | 0.0 |
| PRINTS | PR00702 | Acriflavin resistance protein family signature | IPR001036 | Acriflavin resistance protein | 8 | 32 | 9.0E-100 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.