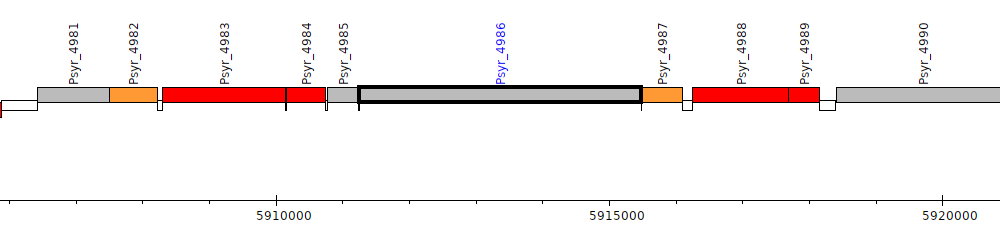
Pseudomonas syringae pv. syringae B728a, Psyr_4986
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF20148 | Domain of unknown function (DUF6531) | IPR045351 | Domain of unknown function DUF6531 | 232 | 305 | 3.1E-21 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 632 | 673 | 0.0082 |
| Gene3D | G3DSA:2.180.10.10 | - | - | - | 823 | 1275 | 4.3E-84 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 676 | 714 | 2.2E-5 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1244 | 1263 | 2.7E-15 |
| PANTHER | PTHR32305 | - | - | - | 365 | 1298 | 9.4E-32 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 859 | 894 | 2.6E-7 |
| Pfam | PF05488 | PAAR motif | IPR008727 | PAAR motif | 73 | 138 | 2.4E-16 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 859 | 894 | 3.1E-5 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1221 | 1241 | 2.7E-15 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 576 | 609 | 3.4E-4 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 527 | 566 | 2.8E-6 |
| Pfam | PF03527 | RHS protein | IPR001826 | RHS protein | 1173 | 1206 | 6.1E-19 |
| NCBIfam | TIGR03696 | JCVI: RHS repeat-associated core domain | IPR022385 | Rhs repeat-associated core | 1200 | 1276 | 1.8E-26 |
| Gene3D | G3DSA:2.60.200.60 | - | - | - | 62 | 148 | 2.3E-22 |
| PRINTS | PR00394 | RHS protein signature | IPR001826 | RHS protein | 1167 | 1187 | 2.7E-15 |
| NCBIfam | TIGR01643 | JCVI: YD repeat (two copies) | IPR006530 | YD repeat | 737 | 777 | 1.0E-4 |
| CDD | cd14742 | PAAR_RHS | - | - | 75 | 148 | 7.78283E-35 |
| Gene3D | G3DSA:3.90.930.1 | - | - | - | 654 | 770 | 9.2E-6 |
| Gene3D | G3DSA:2.180.10.10 | - | - | - | 405 | 624 | 1.1E-22 |
| Pfam | PF05593 | RHS Repeat | IPR031325 | RHS repeat | 527 | 562 | 5.8E-5 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.