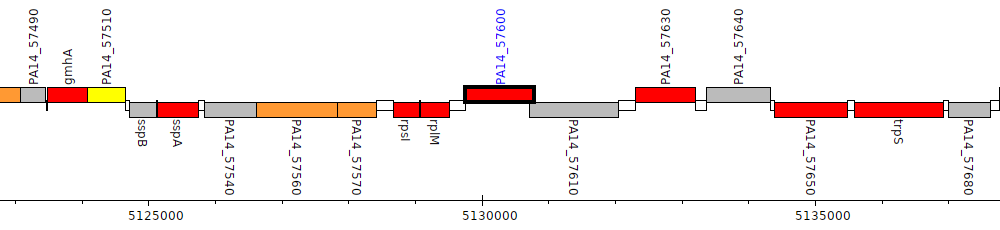
Pseudomonas aeruginosa UCBPP-PA14, PA14_57600
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00069
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF51430 | NAD(P)-linked oxidoreductase | IPR036812 | NADP-dependent oxidoreductase domain superfamily | 1 | 343 | 5.11E-105 |
| Gene3D | G3DSA:3.20.20.100 | - | IPR036812 | NADP-dependent oxidoreductase domain superfamily | 1 | 345 | 3.3E-106 |
| FunFam | G3DSA:3.20.20.100:FF:000005 | NADP(H)-dependent aldo-keto reductase | - | - | 1 | 345 | 0.0 |
| PRINTS | PR00069 | Aldo-keto reductase signature | IPR020471 | Aldo-keto reductase | 116 | 134 | 4.5E-7 |
| CDD | cd19094 | AKR_Tas-like | - | - | 13 | 336 | 0.0 |
| PANTHER | PTHR43364 | NADH-SPECIFIC METHYLGLYOXAL REDUCTASE-RELATED | - | - | 1 | 341 | 5.5E-97 |
| PRINTS | PR00069 | Aldo-keto reductase signature | IPR020471 | Aldo-keto reductase | 163 | 180 | 4.5E-7 |
| Pfam | PF00248 | Aldo/keto reductase family | IPR023210 | NADP-dependent oxidoreductase domain | 16 | 336 | 2.8E-65 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.