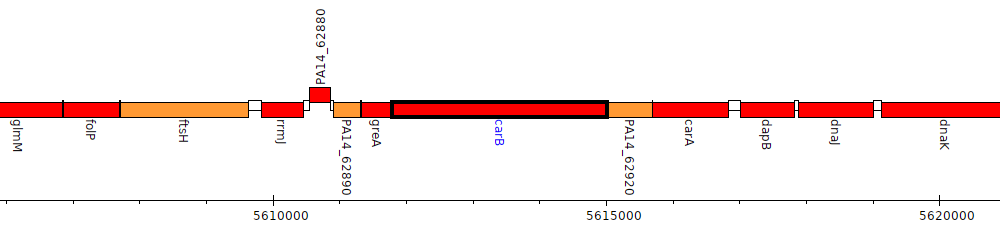
Pseudomonas aeruginosa UCBPP-PA14, PA14_62910 (carB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0071230 | cellular response to amino acid stimulus |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA4756
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
8169201 | |
| Biological Process | GO:0006536 | glutamate metabolic process |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA4756
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
6402363 | |
| Molecular Function | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA4756
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
6402363 | |
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF02786
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006807 | nitrogen compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01369
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pau00250 | Alanine, aspartate and glutamate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau00240 | Pyrimidine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.30.1490.20:FF:000001 | Carbamoyl-phosphate synthase large chain | - | - | 686 | 755 | 1.3E-26 |
| Gene3D | G3DSA:3.30.470.20 | - | - | - | 665 | 935 | 8.0E-120 |
| SUPERFAMILY | SSF56059 | Glutathione synthetase ATP-binding domain-like | - | - | 128 | 401 | 1.94E-109 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 239 | 256 | 4.0E-66 |
| SUPERFAMILY | SSF52335 | Methylglyoxal synthase-like | IPR036914 | Methylglyoxal synthase-like domain superfamily | 938 | 1069 | 1.01E-39 |
| NCBIfam | TIGR01369 | JCVI: carbamoyl-phosphate synthase (glutamine-hydrolyzing) large subunit | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 2 | 1053 | 0.0 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 380 | 398 | 4.0E-66 |
| PANTHER | PTHR11405 | CARBAMOYLTRANSFERASE FAMILY MEMBER | - | - | 6 | 1061 | 0.0 |
| FunFam | G3DSA:1.10.1030.10:FF:000002 | Carbamoyl-phosphate synthase large chain | - | - | 405 | 553 | 1.7E-58 |
| FunFam | G3DSA:3.30.470.20:FF:000007 | Carbamoyl-phosphate synthase large chain | - | - | 119 | 403 | 0.0 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 204 | 223 | 4.0E-66 |
| SUPERFAMILY | SSF52440 | PreATP-grasp domain | IPR016185 | Pre-ATP-grasp domain superfamily | 1 | 127 | 2.01E-49 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 48 | 58 | 4.0E-66 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 297 | 326 | 4.0E-66 |
| Gene3D | G3DSA:1.10.1030.10 | - | IPR036897 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain superfamily | 404 | 553 | 1.2E-54 |
| Gene3D | G3DSA:3.40.50.20 | - | - | - | 1 | 116 | 8.7E-49 |
| FunFam | G3DSA:3.40.50.1380:FF:000004 | Carbamoyl-phosphate synthase large chain | - | - | 936 | 1072 | 2.5E-63 |
| CDD | cd01424 | MGS_CPS_II | IPR033937 | Carbamoyl-phosphate synthase large chain, methylglyoxal synthase-like domain | 942 | 1051 | 9.32562E-50 |
| SUPERFAMILY | SSF52440 | PreATP-grasp domain | IPR016185 | Pre-ATP-grasp domain superfamily | 557 | 675 | 2.3E-49 |
| Pfam | PF02786 | Carbamoyl-phosphate synthase L chain, ATP binding domain | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 673 | 875 | 1.1E-32 |
| Gene3D | G3DSA:3.40.50.1380 | - | IPR036914 | Methylglyoxal synthase-like domain superfamily | 936 | 1072 | 1.6E-42 |
| Pfam | PF02787 | Carbamoyl-phosphate synthetase large chain, oligomerisation domain | IPR005480 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain | 427 | 505 | 1.8E-25 |
| Gene3D | G3DSA:3.30.470.20 | - | - | - | 119 | 403 | 0.0 |
| Pfam | PF02786 | Carbamoyl-phosphate synthase L chain, ATP binding domain | IPR005479 | Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain | 128 | 334 | 1.6E-72 |
| SUPERFAMILY | SSF56059 | Glutathione synthetase ATP-binding domain-like | - | - | 673 | 933 | 6.72E-102 |
| Gene3D | G3DSA:3.40.50.20 | - | - | - | 554 | 662 | 5.6E-51 |
| FunFam | G3DSA:3.30.470.20:FF:000013 | Carbamoyl-phosphate synthase large chain | - | - | 665 | 935 | 0.0 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 19 | 33 | 4.0E-66 |
| PRINTS | PR00098 | Carbamoyl-phosphate synthase protein CPSase domain signature | IPR005483 | Carbamoyl-phosphate synthase large subunit, CPSase domain | 168 | 180 | 4.0E-66 |
| Hamap | MF_01210_B | Carbamoyl-phosphate synthase large chain [carB]. | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 1 | 1062 | 13.125657 |
| FunFam | G3DSA:3.40.50.20:FF:000003 | Carbamoyl-phosphate synthase large chain | - | - | 554 | 662 | 1.2E-63 |
| SUPERFAMILY | SSF48108 | Carbamoyl phosphate synthetase, large subunit connection domain | IPR036897 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain superfamily | 396 | 554 | 6.28E-53 |
| SMART | SM00851 | MGS_2a | IPR011607 | Methylglyoxal synthase-like domain | 955 | 1041 | 2.7E-32 |
| SMART | SM01096 | CPSase_L_D3_2 | IPR005480 | Carbamoyl-phosphate synthetase, large subunit oligomerisation domain | 424 | 547 | 6.4E-75 |
| FunFam | G3DSA:3.40.50.20:FF:000001 | Carbamoyl-phosphate synthase large chain | - | - | 1 | 116 | 2.2E-54 |
| Pfam | PF02142 | MGS-like domain | IPR011607 | Methylglyoxal synthase-like domain | 958 | 1041 | 2.0E-19 |
| Hamap | MF_01210_A | Carbamoyl-phosphate synthase large chain [carB]. | IPR006275 | Carbamoyl-phosphate synthase, large subunit | 5 | 1067 | 13.404976 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.