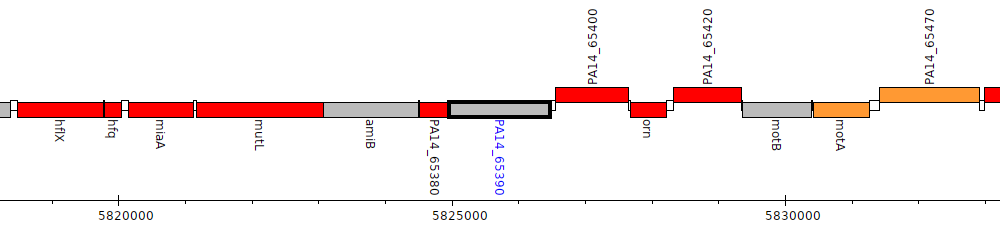
Pseudomonas aeruginosa UCBPP-PA14, PA14_65390
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0052855 | ADP-dependent NAD(P)H-hydrate dehydratase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF017184
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016836 | hydro-lyase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_01965
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| MetaCyc | NADH repair | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF64153 | YjeF N-terminal domain-like | IPR036652 | YjeF N-terminal domain superfamily | 14 | 251 | 1.31E-59 |
| PANTHER | PTHR12592 | ATP-DEPENDENT (S)-NAD(P)H-HYDRATE DEHYDRATASE FAMILY MEMBER | - | - | 86 | 497 | 1.3E-54 |
| NCBIfam | TIGR00196 | JCVI: NAD(P)H-hydrate dehydratase | IPR000631 | ATP/ADP-dependent (S)-NAD(P)H-hydrate dehydratase | 236 | 494 | 4.8E-65 |
| Gene3D | G3DSA:3.40.50.10260 | - | IPR036652 | YjeF N-terminal domain superfamily | 11 | 222 | 1.5E-53 |
| FunFam | G3DSA:3.40.1190.20:FF:000017 | Multifunctional fusion protein | - | - | 226 | 496 | 9.0E-109 |
| Hamap | MF_01965 | ADP-dependent (S)-NAD(P)H-hydrate dehydratase [nnrD]. | IPR000631 | ATP/ADP-dependent (S)-NAD(P)H-hydrate dehydratase | 229 | 499 | 21.26092 |
| NCBIfam | TIGR00197 | JCVI: NAD(P)H-hydrate epimerase | IPR004443 | YjeF N-terminal domain | 16 | 213 | 9.2E-41 |
| CDD | cd01171 | YXKO-related | IPR000631 | ATP/ADP-dependent (S)-NAD(P)H-hydrate dehydratase | 242 | 487 | 1.10763E-88 |
| Hamap | MF_01966 | NAD(P)H-hydrate epimerase [nnrE]. | IPR004443 | YjeF N-terminal domain | 16 | 215 | 22.674129 |
| Pfam | PF03853 | YjeF-related protein N-terminus | IPR004443 | YjeF N-terminal domain | 36 | 194 | 1.7E-39 |
| Pfam | PF01256 | Carbohydrate kinase | IPR000631 | ATP/ADP-dependent (S)-NAD(P)H-hydrate dehydratase | 252 | 487 | 5.7E-60 |
| SUPERFAMILY | SSF53613 | Ribokinase-like | IPR029056 | Ribokinase-like | 233 | 495 | 4.58E-73 |
| Gene3D | G3DSA:3.40.1190.20 | - | IPR029056 | Ribokinase-like | 227 | 497 | 7.9E-84 |
| PIRSF | PIRSF017184 | Nnr | IPR030677 | Bifunctional NAD(P)H-hydrate repair enzyme Nnr | 12 | 498 | 1.4E-127 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.