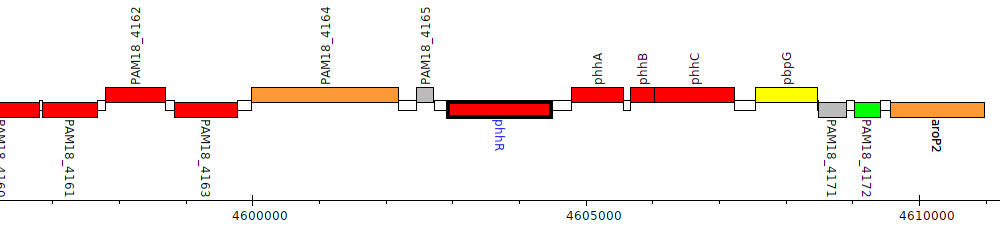
Pseudomonas aeruginosa M18, PAM18_4166 (phhR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006559 | L-phenylalanine catabolic process |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA0873
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
8939433 | |
| Molecular Function | GO:0008134 | transcription factor binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00158
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00158
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SM00382
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR04381
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:PF00158
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| CDD | cd00009 | AAA | - | - | 210 | 374 | 6.47217E-20 |
| SUPERFAMILY | SSF55785 | PYP-like sensor domain (PAS domain) | IPR035965 | PAS domain superfamily | 80 | 138 | 3.66E-9 |
| FunFam | G3DSA:3.30.450.20:FF:000052 | Sigma-54 dependent transcriptional regulator | - | - | 80 | 187 | 1.4E-64 |
| CDD | cd00130 | PAS | IPR000014 | PAS domain | 88 | 145 | 2.29169E-6 |
| NCBIfam | TIGR04381 | JCVI: TyrR family helix-turn-helix domain | IPR030828 | TyrR family, helix-turn-helix domain | 465 | 511 | 5.9E-21 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 223 | 368 | 1.2E-6 |
| SUPERFAMILY | SSF46689 | Homeodomain-like | IPR009057 | Homeobox-like domain superfamily | 455 | 510 | 1.43E-7 |
| PANTHER | PTHR32071 | TRANSCRIPTIONAL REGULATORY PROTEIN | - | - | 150 | 510 | 2.6E-104 |
| FunFam | G3DSA:1.10.10.60:FF:000112 | TyrR family transcriptional regulator | - | - | 455 | 512 | 1.6E-28 |
| Pfam | PF00158 | Sigma-54 interaction domain | IPR002078 | RNA polymerase sigma factor 54 interaction domain | 204 | 370 | 1.8E-58 |
| Gene3D | G3DSA:1.10.8.60 | - | - | - | 376 | 449 | 1.6E-20 |
| Gene3D | G3DSA:3.30.70.260 | - | - | - | 1 | 78 | 9.1E-21 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 203 | 445 | 4.9E-61 |
| SUPERFAMILY | SSF55021 | ACT-like | IPR045865 | ACT-like domain | 2 | 68 | 5.41E-8 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 189 | 375 | 3.2E-58 |
| Gene3D | G3DSA:1.10.10.60 | - | - | - | 458 | 511 | 1.4E-18 |
| Pfam | PF00989 | PAS fold | IPR013767 | PAS fold | 79 | 136 | 1.6E-6 |
| Pfam | PF18024 | Helix-turn-helix domain | IPR030828 | TyrR family, helix-turn-helix domain | 463 | 510 | 8.3E-13 |
| Gene3D | G3DSA:3.30.450.20 | PAS domain | - | - | 80 | 187 | 1.5E-20 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 495 | 515 | - |
| Pfam | PF01842 | ACT domain | IPR002912 | ACT domain | 2 | 60 | 4.6E-7 |
| SMART | SM00091 | pas_2 | IPR000014 | PAS domain | 78 | 144 | 9.6E-9 |
| FunFam | G3DSA:3.40.50.300:FF:000006 | DNA-binding transcriptional regulator NtrC | - | - | 194 | 374 | 1.9E-69 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.