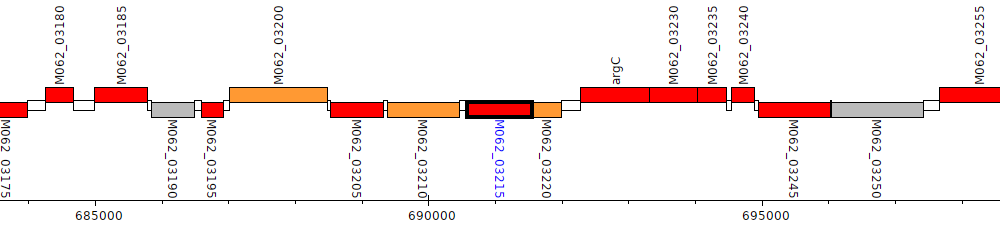
Pseudomonas aeruginosa RP73, M062_03215
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0018580 | nitronate monooxygenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd04730
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | prp00910 | Nitrogen metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR42747 | NITRONATE MONOOXYGENASE-RELATED | - | - | 1 | 84 | 3.8E-88 |
| Coils | Coil | Coil | - | - | 299 | 319 | - |
| CDD | cd04730 | NPD_like | IPR004136 | Nitronate monooxygenase | 11 | 240 | 7.93994E-71 |
| FunFam | G3DSA:3.20.20.70:FF:000210 | 2-nitropropane dioxygenase | - | - | 8 | 312 | 0.0 |
| Pfam | PF03060 | Nitronate monooxygenase | IPR004136 | Nitronate monooxygenase | 89 | 302 | 1.6E-27 |
| SUPERFAMILY | SSF51412 | Inosine monophosphate dehydrogenase (IMPDH) | - | - | 6 | 305 | 8.97E-59 |
| Gene3D | G3DSA:3.20.20.70 | Aldolase class I | IPR013785 | Aldolase-type TIM barrel | 8 | 318 | 2.6E-70 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.