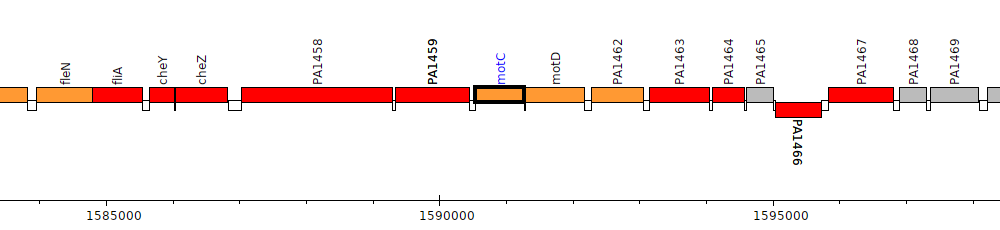
Pseudomonas aeruginosa PAO1, PA1460 (motC)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0071973 | bacterial-type flagellum-dependent cell motility | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
15375113 | Reviewed by curator |
| Biological Process | GO:0071978 | bacterial-type flagellum-dependent swarming motility | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
15629949 | Reviewed by curator |
| Biological Process | GO:0071978 | bacterial-type flagellum-dependent swarming motility |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR30433
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006935 | chemotaxis |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR30433
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02040 | Flagellar assembly | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae02030 | Bacterial chemotaxis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae02020 | Two-component system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Flagella assembly |
ECO:0000037
not_recorded |
|||
| PseudoCAP | Chemotaxis |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF20560 | Motility protein A N-terminal | IPR046786 | Motility protein A, N-terminal | 6 | 69 | 1.9E-7 |
| Pfam | PF01618 | MotA/TolQ/ExbB proton channel family | IPR002898 | MotA/TolQ/ExbB proton channel | 104 | 219 | 4.0E-29 |
| PANTHER | PTHR30433 | CHEMOTAXIS PROTEIN MOTA | IPR047055 | Motility protein A-like | 1 | 245 | 7.6E-78 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.